* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download VIRTUAL COUNTER SCREENING: KINASE INHIBITOR STUDY
Phosphorylation wikipedia , lookup
Signal transduction wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Homology modeling wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
List of types of proteins wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Proteolysis wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
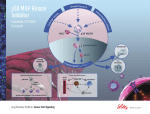
VIRTUAL COUNTER SCREENING: KINASE INHIBITOR STUDY. Daniel N. Santiago, Ashley A. Durand, MinhPhuong Tran, Rachel R. Scheerer, Wayne C. Guida, Wesley H. Brooks, Department of Chemistry, University of South Florida and H. Lee Moffitt Cancer Center & Research Institute, Tampa, Florida 33620. In virtual counter screening (VCS), or inverse docking, a small molecule of interest is docked against a database containing structures of multiple proteins. The VCS approach is potentially useful for measuring (A) drug re-positioning, (B) toxicity, (C) metabolic degradation, (D) lead optimization, and (D) focusing focused libraries. Thus far, we have prepared approximately 1,800 proteins from the Protein Data Bank for docking with GLIDE. Statistics for each protein structure are generated by docking a calibration set of small molecule structures (NCI Diversity Set I). These statistics allow us to determine a protein “hit” for a particular molecule by normalizing protein structures in the library. In a kinase inhibitor case study, VCS matched 70% of 860 data points.