* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Krebs cycle
Metalloprotein wikipedia , lookup
Photosynthesis wikipedia , lookup
Basal metabolic rate wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Light-dependent reactions wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Butyric acid wikipedia , lookup
Biosynthesis wikipedia , lookup
Mitochondrion wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Microbial metabolism wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Electron transport chain wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Phosphorylation wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
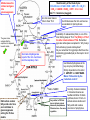
Ferchmin 2016 Index: 1. 2. 3. 4. 5. 6. Pyruvate dehydrogenase complex, structure, cofactors, mechanism, regulation. TCA or Kreb’s cycle, function and regulation. Asymmetric breakdown of a symmetric citrate molecule. “Accounting” of ATP synthesized, from glycolysis to respiratory chain. Murphy’s Law and the missing 10 molecules of ATP per glucose. How to avoid losing your carbons during the exam. Steps shown and enzymes involved. 1 and 2) Pyruvate dehydrogenase (not decarboxylase). 3 and 4) Dihydrolipoyl transacetylase. 5) Dihydrolipoyl dehydrogenase How come NAD++H transfers H to FAD? The –SH reduces the other –S- of lipoic acid The pyruvate dehydrogenase complex is s true multienzymatic complex Electron micrography of the PDC of E. coli. Model of the PDC of E. coli. Red: E1 pyruvate dehydrogenase Yellow: E2 transacetylase Green: E3 dihydrolipoyl dehydrogenase Regulation of the PDC by allosteric and covalent modulation The enzymes involved Coenzymes Description Activators Inhibitors 1) Pyruvate dehydrogenase (E1) Thiamine pyrophosphate (Vitamin B1) Decarboxylation of pyruvate, E1α and E1β, phosphorylation site of PDC. 30 subunits AMP energy GTP energy 2) Lipoate transacetylase (E2 ) Lipoic acid Dehydrogenation of hydroxyethyl and transfer of acetyl to CoA. 60 subunits CoA TCA substrate CoA~Acetyl TCA substrate 3) Dihydrolipoyl dehydrogenase (E3) NAD+ and FAD (Niacin & B2) Regeneration of the lipoate and reduction of NAD+ to NADH. 10 subunits NAD+ Reducing power NADH; Reducing power 1) Pyruvate dehydrogenase kinase inactivates the PDC Complex by phosphorylation of three serines on E1α. PRODUCTS ATP Acetyl CoA NADH SUBSTRATES ADP; pyruvate; CoA; NAD+ 2) Pyruvate dehydrogenase phosphatase removes the inhibition imposed by the phosphorylation through hydrolysis of the Pi. Ca2+ insulin NADH The regulatory subunits of the PDC are intrinsic to the complex Ca2+ in muscle & insulin in liver Insulin (means abundance of glucose) disinhibits the PDC and reroutes pyruvate from gluconeogenesis to lipogenesis Malate leaves the mitosol and goes into gluconeogenesis Stoichiometry of the Krebs Cycle: CH3-CO-CoA + 3 NAD+ FAD + GDP + Pi + 2 H2O 2 CO2 + 3 NADH + FADH2 + GTP + 2 H+ + CoA ΔG’°=1.1 kcal/mole (sluggish) Ac~CoA never leaves mito to favor TCA The green arrow indicates that the equilibrium is displaced towards malate. Succinate dehydrogenase is part of the mito. membrane and the respiratory chain but citrate leaves the mito. and serves as substrate for lipid synthesis Availability of oxaloacetate (OAA) is one of the main limiting steps of TCA. The [OAA] is 1/10 of the other intermediates of TCA. Remember pyruvate carboxylase is anaplerotic. Why AcetylCoA activates pyruvate carboxylase? Do you remember from glycolysis that the active metabolite (glyceraldehyde) is often kept in short supply? Isocitrate dehydrogenase is the key enzyme (committed step). Strictly dependent on the ration of ADP/ATP and NAD+/NADH. Makes TCA aerobic by the “substrate control”. Odd carbon number fatty acids enter here and contribute to gluconeogenesis exiting the TCA as malate. Very similar to PDC but has no intrinsic protein kinases & phosphatases. Otherwise has ~ the same regulation Succinyl~CoA accumulates in mitosol and serves as feedback inhibitor of citrate synthase, donor of CoA~ to activate ketone bodies and fatty acids and is also a precursor of porphyrines. Aconitase is inhibited by fluoroacetate an enzyme activated inhibitor (often called suicidal enzyme inhibitor). Malonic (HOCO-CH2-COOH) acid is the archetypal example of a competitive inhibitor: it acts against succinate dehydrogenase (complex II) in the respiratory electron transport chain. Odd carbon acids converts into propionylCoA which cannot directly enter either beta oxidation or the citric acid cycles. Instead it is carboxylated to D-methylmalonyl-CoA, which is isomerized to L-methylmalonylCoA. A vitamin B12-dependent enzyme catalyzes rearrangement of Lmethylmalonyl-CoA to succinyl-CoA, which is an intermediate of the citric acid cycle. The cycle is totally aerobic because of substrate (availability) control. Notice however, that O2 is not actually present. The * indicate the requirement of an oxidized NAD+ or FAD for the TCA to proceed. The II indicate the requirement of ADP for the cycle to proceed. The yield of ATP per NADH+H+ depends on the shuttle used. The next slide is for you to review after you had the lecture about oxidative/phosphorylation. The objective is to account for all the moles of ATP produced per mole of glucose metabolized to CO2 and H2O. The purpose is to get familiarized with the intricacy of interactions among metabolic pathways ATP YIELD FROM THE COMPLETE OXIDATION OF GLUCOSE Reaction sequence ATP yield per glucose Glycolysis: glucose into pyruvate (in the cytosol) Phosphorylation of glucose Phosphorylation of fructose 6-phosphate Dephosphorylation of 2 molecules of 1,3-DPG Dephosphorylation of 2 molecules of phosphoenolpyruvate -1 -1 +2 +2 2 NADH are formed in the oxidation of 2 molecules of * glyceraldehyde 3-phosphate (Shuttle to mitochondria and oxidative phosphorylation) Conversion of pyruvate into acetyl CoA (inside mitochondria) 2 NADH are formed ** Citric acid cycle (inside mitochondria) 2 molecules of guanosine triphosphate are formed from 2 molecules of succinyl CoA 6 NADH are formed in the oxidation of 2 molecules of each isocitrate, α-ketoglutarate, and malate 2 FADH2 are formed in the oxidation of 2 molecules of succinate +2 Oxidative phosphorylation (inside mitochondria) 2 NADH formed in glycolysis; each yields 2 ATP (Assuming transport of NADH by the glycerol phosphate shuttle) 2 NADH formed in the oxidative decarboxylation of pyruvate: each yields 3 ATP 2 FADH2 formed in the citric acid cycle; each yields 2 ATP 6 NADH formed in the citric acid cycle; each yields 3 ATP +4 * +6 ** +4 +18 +36 or 38 These 36 ATP could be 38 if the malate aspartate shuttle is used. However, in real life, the mitochondrion uses part of its H+ gradient for “housekeeping” and the approximate yield of ATP/mol of glucose is roughly 24.