* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download How does Pol II
Deoxyribozyme wikipedia , lookup
Magnesium transporter wikipedia , lookup
Non-coding RNA wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein moonlighting wikipedia , lookup
Molecular evolution wikipedia , lookup
Promoter (genetics) wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Polyadenylation wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Interactome wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene regulatory network wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression wikipedia , lookup
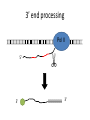
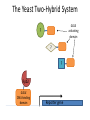
Hannah Baer Hawley Lab DNA Pol II RNA ribosome protein How does Pol II “know” when to terminate transcription? Termination is linked to 3’ end processing. 3’ end processing Pol II 5’ 5’ AAAAAAA 3’ Anti-terminator vs. torpedo models Pol II Anti-terminator: recognition of the poly(A) sequence causes a conformational change Torpedo: transcript cleavage allows degradation to occur How is termination linked to polyadenylation? Termination-defective mutants What is the nature of the defects? What interactions are there that could have been disrupted by the mutations? The Yeast Two-Hybrid System GAL4 activating domain ? ? ? Rpb2 GAL4 DNA-binding domain Reporter gene Experiments • Screen 1 million transformants from each of three libraries • Test for expression of all three reporter genes: ADE2, HIS3, and lacZ • Test any “positive” preys against four mutant baits Assays for reporter gene expression left = HIS3; right = lacZ Results • Fourteen colonies expressed all three reporters, but only four were real proteins – RSC2, CIN5, YRF1, and YLR035C-A • None of the preys interacted with any of the mutant baits • Two TFIIF prey constructs failed to interact with the mutant or wild-type baits Summary • Three million transformants screened; three interactions found • When these were tested against the mutant baits, results were inconclusive Verify that the fusion proteins are being expressed Screen more from the three libraries Characterize the remaining 36 mutants Acknowledgments • • • • Diane Hawley Susan Strachan Brittney Lilly SPUR