* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download C303, Teaching Building 2015/09 Genetic Susceptibility(易感性)
Biology and sexual orientation wikipedia , lookup
Pathogenomics wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
Tay–Sachs disease wikipedia , lookup
Human genome wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic testing wikipedia , lookup
Genetic drift wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome evolution wikipedia , lookup
Nutriepigenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Medical genetics wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Designer baby wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Heritability of IQ wikipedia , lookup
Genome (book) wikipedia , lookup
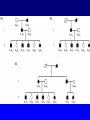
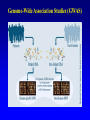
复杂疾病的遗传学 张咸宁 [email protected] Tel:13105819271; 88208367 Office: C303, Teaching Building 2015/09 Genetic Susceptibility(易感性) • An inherited predisposition to a disease or disorder which is not due to a single-gene cause and is usually the result of a complex interaction of the effects of multiple different genes, i.e. polygenic inheritance. 遗传因素和环境因素在疾病中的作用 健康 / 疾病 = 基因 + 环境因素 单基因疾病 致病基因 多因子疾病 外伤 易感基因 环境 基因 血友病 大肠癌 老年痴呆 中风 心血管疾病 肺癌 侏儒 乳腺癌 糖尿病 皮肤癌 哮喘 交通意外 Why study the genetics of common diseases? • Understanding the underlying genetics will lead to understanding the causes, which may lead to better and more specific therapies • Identification of the responsible genes helps to identify those who are at risk in families and in the population, allowing individualized health assessment and targeted prevention How to Determine the Genetic Components of Complex Diseases? • Family, twin and adoption studies(家系、双生子 和寄养子分析) • Segregation analysis(分离分析) • Linkage analysis(连锁分析) • Association studies and linkage disequilibrium (关联研究和连锁不平衡分析) • Identification of DNA sequence variants conferring susceptibility(鉴定易感基因的突变) • Trait: Any detectable phenotypic property or character. • Qualitative trait(质量性状): A genetic disease trait that either present or absent. The pattern of inheritance for a qualitative trait is typically monogenetic, which means that the trait is only influenced by a single gene. • Quantitative trait(数量性状) : are measurable characteristics such as height, blood pressure, serum cholesterol, and body mass index. A quantitative trait shows continued variation under the influence of many different genes. Quantitative trait:Normal distribution • • • • • • • Height Weight Shape Color Blood pressure metabolic activity reproductive rate Polygenes: small-but-equal effect QTL (quantitative trait loci) Successive Approximations to a Gaussian Distribution(逐次逼近正态分布): QTL Liability(易患性) • A concept used in disorders which are multifactorially determined to take into account all possible causative factors. threshold: 阈值 Familial aggregation(家族聚集性): Affected individuals tend to cluster in families. prevalence(患病率) of the disease in a relative “r” of an affected person lr= --------------------------------------------------population prevalence of the disease • The higher the familial aggregation, the larger the lr. • If lr = 1, then the relative is at no greater risk than anyone in the general population. Twin studies suffer from many limitations • Monozygotic (MZ。单卵双生子) twins are genetically identical clones and should always be concordant (both the same.一致性) for any genetically determined character. • Dizygotic (DZ。两卵双生子) twins share half their genes on average, the same as any pair of sibs(同胞对). Genetic Differences between Identical Twins • All individuals, even MZs, differ in: – their repertoire(库)of antibodies and T-cell receptors (because of epigenetic rearrangements and somatic cell mutations); – somatic mutations in general – the numbers of mitochondrial DNA molecules (epigenetic partitioning); – the pattern of X inactivation, if female. Genetic analysis of quantitative traits • Correlation(相关): • Heritability(遗传率): Correlation is a statistical measure of the degree of association of variable phenomena (a measure of the degree of resemblance or relationship between 2 parameters). Coefficient of correlation (r) • Positive correlation(正相关): r>1 • No correlation: r=0 • Negative correlation(负相关): r<1 Heritability (h2)遗传率: The proportion of the total variation of a character attributable to genetic as opposed to environment factors. CMZ -- CDZ • h2 = ----------------------100 -- CDZ • If cMZ >> cDZ then h2 is high (approaches 1) • If cMZ = cDZ then h2 is low (approaches 0) [c = concordance(一致性)] Heritability (h2) for Various Diseases Trait or Disease Concordance Rate MZ twins DZ twins Heritability Alcoholism Autism Cleft lip/palate Diabetes, type 1 Diabetes, type 2 Measles Schizophrenia 0.6 0.92 0.38 0.35-0.5 0.7-0.9 0.95 0.47 0.6 >1 0.6 0.6-0.8 0.9-1.0 0.16 0.7 0.3 0.0 0.08 0.05-0.1 0.25-0.4 0.87 0.12 Mendelian Forms of Common Complex Diseases Common Disease Mendelian Subtype Atherosclerosis Breast cancer Familial hypercholesterolemia Familial breast/ovarian cancer Amyotrophic lateral Familial ALS Sclerosis Parkinson disease Alzheimer disease Hypertension Involved Gene LDL receptor (LDLR) BRCA1, BRCA2 Superoxide dismutase (SOD1) Familial Parkinson disease -synuclein Familial AD PS1, PS2, APP Liddle syndrome Renal sodium channel (SCNN1B) Spectrum of Complexity for Common Diseases: From “Simple” to Complex Alzheimer disease (AD) • Complex genetic contributions to AD may come from: – One or more incompletely penetrant genes that act independently; – Multiple interacting genes; or – Combination of genetic and environmental factors Spectrum of Complexity for Common Diseases: From “Simple” to Complex Alzheimer disease (AD) • Familial AD – Approximately 10% of patients have a monogenic form of AD with highly penetrant, age-related, autosomal dominant inheritance – Presents earlier than typical AD: as early as 3rd decade (20s) compared with 7th-9th decades for typical AD – Three genes: PS1, PS2, APP Even “Sporadic” AD May Have a Genetic Component • Apolipoprotein E (APOE) – Protein component of LDL particle – Constituent of amyloid plaques in AD – Three alleles: 2, 3, 4 • • • • 4/4: >90% show AD by age 80 2/3: <10% show AD by age 80 4/- : 25-50% show AD by age 80 Environmental factors also involved – Association between presence of 4 allele and AD following head trauma is seen in professional boxers Genetic Testing for APOE Genotypes • Testing asymptomatic(无症状) individuals for 4 remains controversial(争论) – Poor predictive value(预测值) – No effective therapeutic intervention(治疗干预措 施) available to prevent onset(发病) • Available through direct-to-consumer marketing(面向消费者的市场) CNVs are common in all genomes surveyed … • Blue = pathogenic • Red = deletion • Green = duplication Genetic mapping of complex traits • Linkage analysis(连锁分析) • Association studies(关联研究) Genetic mapping of complex traits Linkage analysis: genome scan(全基因组扫描) which analyzes the disease pedigrees using hundreds of polymorphic markers (SNP) throughout the entire genome. L (θ) • Lods (log odds score): Z(θ) = log[------------ ] L (1/2) Lod score (z) • A measure of the likelihood of genetic linkage between loci.The log (base 10) of the odds that the loci are linked (with recombination θ) rather than unlinked. • For mendelian characters a lod score greater than +3 is evidence of linkage;one that is less than –2 is evidence against linkage. Identity by State (IBS) and Identity by Descent (IBD) • Both sib pairs share allele A1. The first sib pair have two independent copies of A1 (IBS but not IBD); the second sib pair share copies of the same paternal A1 allele (IBD). The difference is only apparent if the parental genotypes are known. Sib Pair Analysis (A)By random segregation sib pairs share 0, 1 or 2 parental haplotypes 1/4, 1/2 and 1/4 of the time, respectively. (B) Pairs of sibs who are both affected by a dominant condition share one or two parental haplotypes for the relevant chromosomal segment. (C) Pairs of sibs who are both affected by a recessive condition share both parental haplotypes for the relevant chromosomal segment. Affected sibling-pair(患病同胞对) analysis Suggested Criteria for Reporting Linkage Category of linkage Expected No. of occurrences by chances in a whole genome scan Range of approximate p value Range of approximate lod scores Suggestive 1 7 x 10-4 – 3 x 10-5 2.2 - 3.5 Significant 0.05 2 x 10-5 – 4 x 10-7 3.6 - 5.3 Highly significant 0.001 <3 x 10-7 >5.4 Confirmed 0.01 in a search of a candidate region that gave significant linkage in a previous independent study Lod score: 3.6 for IBD testing of affected sib pairs, 4.0 for IBS •Given that the loci are truly linked, with recombination fraction q, the likelihood of a meiosis being nonrecombinant is 1 - θ and the likelihood of it being recombinant is θ. •If the loci are in fact unlinked, the likelihood of a meiosis being either recombinant or nonrecombinant is 1/2. Family A There are five recombinants and one nonrecombinant. The overall likelihood, given linkage, is (1 - θ)5. θ The likelihood given no linkage is (1/2)6 The likelihood ratio is (1 - θ)5. θ / (1/2)6 The lod score, Z, is the logarithm of the likelihood ratio. Family B II1 is phase-unknown. If she inherited A1 with the disease, there are five nonrecombinants and one recombinant. If she inherited A2 with the disease, there are five recombinants and one nonrecombinant. The overall likelihood is 1/2 [(1 - θ)5. θ / (1/2)6] + 1/2 [(1 - θ). θ 5 / (1/2)6]. This allows for either possible phase, with equal prior probability. The lod score, Z, is the logarithm of the likelihood ratio. Family C At this point nonmasochists turn to the computer. Genome-wide scanning: autozygosity mapping Positional cloning(定位克隆) • Disease mapping - chromosome deletion mutation - linkage analysis • Fine mapping • Candidate genes - pathogenic mutation screening - functional assay Another method combines genome scanning and the use of animal models Population Associations • The low success rate of linkage studies for cx traits in the 1990s suggested that many of the susceptibility factors must be relatively weak, highly heterogeneous, or both. • Rather than studying affected relatives, association studies seek populationwide associations between a particular condition and a particular allele or haplotype somewhere in the genome. Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996; 273:1516. Population Associations • Association is simply a statistical statement about the cooccurrence of alleles or phenotypes. HLA-DR4, 36% UK / 78%, rheumatoid arthritis • A population association can have many possible causes, not all of which are genetic. Direct causation: An epistatic effect: Population stratification: HLA*A1 and Type I error: false positives Linkage disequilibrium (LD): Association is quite different from linkage, except where the family and the population merge • Linkage is a specific genetic relationship between loci (physical sites on the cs) • Association is a relation between specific alleles and/or phenotypes. • Linkage does not of itself produce any association in the general population. Linkage creates association within families, but not between unrelated people. Genome-Wide Association Studies (GWAS) Association studies depend on linkage disequilibrium (LD) • The occurrence together of 2 or more alleles at closely linked loci more frequently than would be expected by chance. → allelic association • EMLD (http://request.mdacc.tmc.edu/qhuang/ Software/pub.htm) • D’:0(no LD)~±1 (complete association) SNP: A change in which a single base in the DNA differs from the usual base at that position. Millions of SNP's have been cataloged in the human genome. Some SNPs such that which causes sickle cell are responsible for disease. Other SNPs are normal variations in the genome. Haplotype • A group of alleles in coupling at closely linked loci, usually inherited as a unit. Haplotype(单倍型) Tag SNPs(标记SNP) • A select, minimal subset of all the SNPs in a genomic region, chosen because they are in linkage disequilibrium with one another in the population. Tag SNPs are useful because they form a minimum set of SNPs whose alleles constitute haplotypes capable of representing all the common haplotypes in that region. HapMap: A set of haplotypes, defined by tag SNPs, distributed throughout the genome, used for association studies. Measures of LD(连锁不平衡) • If 2 loci have alleles A, a and B, b with frequencies pA, pa, pB and pb → 4 possible haplotype: AB, Ab, aB and ab; pAB, pAb, paB and pab. • If no LD, pAB= pApB and so on. The degree of departure, D = pABpab – pAbpaB. • D’=(pAB - pApB )/Dmax (the maximum value of ∣pAB - pApB∣possible with the given allele frequencies) • △2=(pAB - pApB)2/(pApapBpb) A new generation of genomewide association studies (GWAS) has finnaly broken the logjam in cx dis research?? Linkage and Association: Complementary Techniques • Linkage operates over a long chromosomal range, scan the entire genome in a few hundred tests – 250 ASP, 300 markers = 1.5-3 x 105 tests – candidate regions impracticably large for positional cloning • LD short range phenomenon – TDT, 300 trios, 25 kb LD = 108 tests – Need to focus on predetermined candidate regions by animal models, known gene, or linkage studies Difficulties in Identification of DNA Sequence Variants Conferring Susceptibility to Cx diseases • No single gene mutation is necessary or sufficient to cause the disease – even a true susceptibility allele will be found in some controls and be absent from some patients. – The main determinants of susceptibility may be different in different populations • No genetic way to identify true determinant among a set of alleles in a strong LD • The genetic variants causing susceptibility to common diseases may not be obvious mutations – May be SNPs in noncoding regions have small effect on promoter activity, RNA splicing or mRNA stability. How to Determine the Genetic Components of Complex Diseases? • • • • Family, twin and adoption studies Segregation analysis Linkage analysis Association studies and linkage disequilibrium • Identification of DNA sequence variants conferring susceptibility Characteristics of Inheritance of Complex Diseases 1. Diseases with complex inheritance are not single-gene disorders and do not demonstrate a simple Mendelian pattern of inheritance. 2. Diseases with complex inheritance demonstrate familial aggregation, because relatives of an affected individual are not likely to have disease-predisposing alleles in common with the affected person than are unrelated individuals. 3. Pairs of relatives who share disease-predisposing genotypes at relevant loci may still be discordant for phenotype(show lack of penetrance) because of the crucial role of nongenetic factors in disease causation. The most extreme examples of lack of penetrance despite identical genotypes are discordant MZ. 4. The disease is more common among the close relatives of the proband and becomes less common in relatives who are less closely related. Greater concordance for disease is expected among MZ verse DZ.

























































![Department of Health Informatics Telephone: [973] 972](http://s1.studyres.com/store/data/004679878_1-03eb978d1f17f67290cf7a537be7e13d-150x150.png)