* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download posterexample1
Genomic imprinting wikipedia , lookup
Gene nomenclature wikipedia , lookup
RNA interference wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genome evolution wikipedia , lookup
Point mutation wikipedia , lookup
Nicotinic acid adenine dinucleotide phosphate wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Designer baby wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Protein moonlighting wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression programming wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
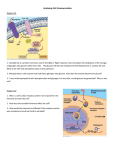
The atToc159 receptor complex represents a critical point in the biosynthesis of jasmonic acid Author and co-authors: Danielle Sprout, Kayla Duffield, Samantha Workman Faculty Advisor: Dr. Afitlhile Department of Biological Sciences Objective We evaluated the ability of ppi2 mutant to accumulate mRNA that encode enzymes that function in the JA pathway. We also measured transcript levels of the JA responsive gene, PDF 1.2 and JAZ1, which encodes a protein that represses JA responsive genes. Hypothesis The atToc159 receptor complex is a major route in the import of LOX-2, AOS and AOC, which are enzymes that function in the initial stages of the JA pathway. We expect the wounded mutant to accumulate low levels of mRNA that encode LOX-2, AOS, OPR-3, and the JA responsive gene, PDF1.2. 6 Wt, uw Wt, w Mt, uw Mt, w 5 Relative fold change 4 3 2 1 0 LOX-2 heterozygous ppi2 AOS OPR-3 Figure 1. Expression of LOX-2, AOS and OPR3 genes in the unwounded and wounded leaves of ppi2 mutant and wild type plants. Data represent the mean ± SEM of 4 replicates. 5 Discussion In the wounded wild type, the expression of LOX-2, AOS, OPR-3, and the JA responsive gene PDF 1.2 was upregulated. Expression of JAZ1 was comparable between the unwounded and wounded tissue, which suggests that the synthesis and degradation of JAZ1 protein was tightly regulated. However, in the wounded ppi2 mutant the expression of LOX-2, AOS, and OPR-3 was suppressed. Interestingly, in ppi2 transcripts for the jasmonate responsive gene, PDF1.2 did not accumulate above the control level. This observation suggests that in the wounded mutant, JA or JA-Ile did not accumulate to levels high enough to induce the expression of PDF1.2 gene. The data indicate that in the absence of Toc159 receptor, enzymes that function in the initial stages of the JA pathway were not efficiently imported into the chloroplasts. We conclude that atToc159 receptor is required in the import of enzymes that function in the JA pathway. Acknowledgments Relative fold change Introduction When plants are eaten by insect herbivores or wounded mechanically, the fatty acid linolenic acid (18:3) is metabolized to produce the plant hormone, jasmonic acid (JA), which accumulates to high levels in wounded tissues. The JA pathway is initiated in the chloroplasts and completed in the peroxisomes. JA is then exported to the cytoplasm where it is conjugated to isoleucine to form JA-Ile. The latter binds to its receptor, which induce signals that turn on an array of plant defense genes, including plant defensin, PDF1.2. Enzymes that function in the JA pathway are encoded by genes that are localized within the nucleus. Lipoxygenase-2 (LOX-2), allene oxide synthase (AOS), and allene oxide cyclase (AOC) are synthesized in the cytoplasm and imported into the chloroplasts, while OPR-3 is imported into the peroxisomes. Proteins that are synthesized in the cytoplasm are recognized by receptors in membranes of the chloroplasts and peroxisomes. Receptors on the outer membrane of the chloroplast include atToc159, atToc132, and atToc120. These receptors are found in the same complex with Toc33/34 and Toc75; the latter is a protein channel. Mutation in atToc159 receptor yields plants with an albino phenotype, and are called plastid protein import deficiency or ppi2. Wt, uw Wt, w Mt, uw 4 3 2 1 0 PDF1.2 JAZ1 Figure 2. Expression of PDF1.2 and JAZ1 genes in the unwounded and wounded leaves of ppi2 mutant and wild type plants. Data represent the mean ± SEM of 4 replicates. LOX-2 1 2 3 AOS 4 1 2 1 2 JAZ1 1 2 OPR-3 3 4 3 4 1 2 3 4 UCE 3 4 Dr. Sue Musser, for assistance in the analysis of the qPCR data (WIU). Dr. Matthew Smith for the gift of the ppi2 seeds (Canada). Figure 3. RT-PCR analysis of the expression of LOX-2, AOS, OPR-3 and JAZ-1. Ubiquitin-conjugating enzyme (UCE) is the internal control. 1: unwounded, wild type. 2: wounded, wild type. 3: unwounded, ppi2. 4: wounded, ppi2.