* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download CHAPTER 16 Advanced Gene Mapping in Eukaryotes
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Oncogenomics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Point mutation wikipedia , lookup
Public health genomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genome evolution wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Minimal genome wikipedia , lookup
Genomic imprinting wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Neocentromere wikipedia , lookup
Quantitative trait locus wikipedia , lookup
X-inactivation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression profiling wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
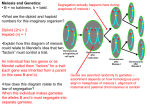
Peter J. Russell A molecular Approach 2nd Edition CHAPTER 16 Advanced Gene Mapping In Eukaryotes edited by Yue-Wen Wang Ph. D. Dept. of Agronomy,台大農藝系 NTU 遺傳學 601 20000 Chapter 13 slide 1 Tetrad Analysis in Certain Haploid Eukaryotes 1. In some haploid eukaryotic organisms (fungi or single-celled algae) products of a single meiosis, the meiotic tetrad, are contained within one structure. Tetrad analysis provides insight into meiotic events. 2. In haploid organisms, the phenotype correlates directly with the genotype of each member of the tetrad (no dominance or recessiveness occurs). 3. Life cycles of organisms typically used in tetrad analysis: a. Saccharomyces cerevisiae (baker’s yeast), has two mating types, MATa and MATα (Figure 16.1). i. Asexual reproduction occurs mitotically (vegetative life cycle) in the haploid yeast. ii. Sexual reproduction, fusion of a haploid a cell with a haploid α one, produces a diploid cell (a/α) that also reproduces mitotically, giving rise to identical diploid cells. iii. Diploid cells sporulate by meiosis, producing four haploid ascospores contained in an ascus. Of the ascospores, two will be type a and two type a. In yeast they are unordered tetrads, arranged randomly in the ascus (Figure 13.16). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 2 Fig. 16.1 Life cycle of the yeast Saccharomyces cerevisiae Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 3 b. Chiamydomonas reinhardtii is a single-celled green algae with haploid vegetative cells and two mating types. i. Nitrogen limitation causes the cells to become gametes, and the two opposite mating types (mt+ and mt-) fuse to produce a zygote. ii. Meiosis of the zygote produces an unordered tetrad of haploid cells, two of type mt+ and two mt-. iii. Mitosis of each haploid cell results in new haploid algae cells. c. Neurospora crassa is similar, but its ascospores are arranged in an ordered tetrad. i. The ordered tetrad reflects the orientation of the four chromatids of the tetrad at the metaphase plate in meiosis I. Spores can be isolated in order or randomly. ii. The ascus contains eight spores, because each haploid cell duplicates by mitosis before spore maturation. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 4 Using Random-Spore Analysis to Map Genes in Haploid Eukaryotes 1. In these organisms, three-point crosses have been used effectively for mapping. Haploidy simplifies interpretation of the results. 2. Analysis of random-spore data is the same as for diploid eukaryotes. It is used for determining linkage and constructing genetic maps (Figure 13.17). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 5 Calculating Gene-Centromere Distance in Organisms with Ordered Tetrads 1. Neurospora’s eight spores represent the result of meiotic division followed by mitosis, and are considered as four pairs. Their order reflects the orientation of the chromatids at metaphase I, allowing the distance between genes and centromere to be mapped (Figure 16.2). 2. Centromeres separate just before the second meiotic division, and so spores in the top of the ascus have the centromere from one parent, while those below have the other parent’s centromere. 3. In this example, mating type (A and a) is one locus, and the centromere is another. a. If no crossover occurs between them, they show first-division segregation. After meiosis I, both copies of A are at one pole and both copies of a at the other. The final result is a 4 : 4 segregation in the ascus. b. Single crossover shows second-division segregation. A and a are each being present in two nuclear areas until the second division, and their pattern of gene segregation depends on the chromatids involved. Both patterns are distinguishable from the 4:4 seen in first-division segregation: i. A 2 : 2 : 2 :2 ratio results from AAaaAAaa and aaAAaaAA. ii. A 2 : 4 : 2 ratio results from AAaaaaAA and aaAAAAaa. c. The distance from the gene of interest (here the mating type locus) to the centromere is the percentage of second-division tetrads. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 6 Fig. 16.2a Determination of gene-centromere distance of the mating-type locus in Neurospora Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 7 Fig. 16.2b Determination of gene-centromere distance of the mating-type locus in Neurospora Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 8 Using Tetrad Analysis to Map Two Linked Genes 1. In a two-point cross, mating produces a diploid heterozygous for both genes, and then meiosis makes haploid spores. In the fungi (Saccharomyces and Neurospora), the spores of the tetrad are micromanipulated for separate germination and analysis. 2. In the cross a+ b+/a b, in which a and b are linked, three different tetrad types can result (Figure 16.3): a. Parental-ditype (PD) tetrad has only the two parental types (a+ b+ and a b). A PD tetrad results either if no crossing-over occurs between the two genes, or if a double crossover involving the same two chromatids occurs. b. Tetratype (T) has two parentals (a+ b+ and a b) and two recombinants (a+ b and a b+). A T tetrad results either from a single crossover between the two genes, or if a double crossover involving three chromatids occurs. c. Non-parental-ditype (NPD) has only recombinants (a+ b and a b+). A NPD tetrad results from a double crossover that involves all four chromatids. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 9 Fig. 16.3 Three types of tetrads Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 10 3. Segregation of alleles is 2:2. Rarely, 3:1 or 1:3 ratios are seen, due to gene conversion (Box 13.2). 4. For genes on different chromosomes, crossover is not involved. PD and NPD tetrads are produced with equal frequency; and no T tetrads are expected (Figure 16.4). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 11 Box Fig. 13.2 Gene conversion by mismatch repair at two sites Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 12 Fig. 16.4 Origin of tetrad types for a cross in which the two genes are located on different chromosomes Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 13 5. When genes are linked (Figure 16.9): a. A single crossover produces a T tetrad. b. Double crossovers vary depending on the strands involved: i. If the same two chromatids are involved in both crossovers, a PD tetrad results. ii. If three chromatids are involved, a T tetrad results. iii. If all four chromatids are involved, an NPD tetrad is produced. c. Genes are considered linked if the PD frequency is far greater than the NPD frequency. d. Genetic distance between the genes correlates with the recombination frequency; and they are mapped accordingly, e. For crosses involving more than two genes, they are considered in pairs, and mapped two at a time relative to each other. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 14 Fig. 16.5 a-c Origin of tetrad types for a cross in which both genes are located on the same chromosome Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 15 Fig. 16.5 d, e Origin of tetrad types for a cross in which both genes are located on the same chromosome Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 16 Mitotic Recombination Discovery of Mitotic Recombination 1. Mitotic crossing-over (mitotic recombination) produces a progeny cell with different gene combinations than the diploid parental cell has. a. Mitotic crossing-over occurs at a stage similar to the 4-strand stage of meiosis. b. This stage is rare, so mitotic crossover is much less frequent than meiotic crossover. 2. Crossing-over during mitosis was first observed by Stern (1936) in Drosophila (Figure 16.6). a. The alleles involved are sex-linked and recessive to wild-type: i. y produces yellow body color instead of wild-type grey. ii. sn produces short, twisty bristles (“singed”) rather than the wild-type long, curved ones. Bristles follow body color (y+/ - are black, and y / y are yellow). b. Female progeny from the cross y+ sn / y+ sn X y sn+ / Y generally have the wild-type phenotype of grey bodies and normal bristles, corresponding to their genotype (y+ sn / y sn+). But exceptions were seen: i. Some flies had patches of yellow and/or singed bristles. This could be explained by nondisjunction or chromosomal loss. ii. Other flies had twin spots, adjacent regions of bristles, one yellow and the other singed, a mosaic phenotype. The spots are reciprocal products of the same genetic event, a mitotic crossing over. c. Mitotic crossover occurred either between the centromere and the sn locus, or between the sn and the y locus (Figure 16.7). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 17 Fig. 16.6 Body surface phenotype segregation in a Drosophila strain Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 18 Fig. 16.7 Production of the twin spot and single yellow spot shown in Figure 16.6 by mitotic crossing-over Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 19 3. Mitotic crossing-over makes all genes distal to the crossover point homozygous if the chromatids align properly at the metaphase plate. a. Proper alignment occurs one half of the time. b. Homozygosity applies only to genes on the same chromosome arm, with no effect on genes of the other arm. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 20 Mitotic Recombination in the Fungus Aspergillus nidulans 1. Meiotic recombination analysis is not used with Aspergillus because the fungus selfs. It is used for studies of mitotic recombination because: a. Its asexual spores are uninucleate. b. Phenotype of each asexual spore is controlled by its nucleus. c. Two haploid strains can be fused by mixing. 2. Heterozygous strains are constructed for study by fusing two haploid strains of different genotype, resulting in a heterokaryon (mycelium with two nuclear types that coexist and divide mitotically). 3. Rarely, haploid nuclei of the heterokaryon fuse to form a diploid nucleus (diploidization) that will form diploid spores. Diploid spores are larger than haploid spores, and may be isolated for study. 4. An example is given in Figure 16.8: a. Allele designations are as follows: i. w (with either y or y+) produces white colonies, w+y produces yellow, and w+y+ produces green. ii. ad cells require adenine in the growth medium, ad+ do not. iii. pro cells require proline in the growth medium, pro+ do not. iv. paba cells require para-aminobenzoic acid (PABA) in the growth medium, paba+ do not. 台大農藝系 v. bi cells require biotin in the growth medium, bi+ do not.遺傳學 601 20000 Chapter 13 slide 21 Fig. 16.8 Photo of a green diploid Aspergillus colony, genotype w ad+ pro paba+ y+ bi/w+ ad pro+ paba y bi+ with a yellow and white sector. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 22 b. In this example, strain 1 is crossed with strain 2: i. Strain 1 is wad+propaba+y+bi ii. Strain 2 is w+adpro+pabaybi+ iii. The diploid strain produced will have wild-type alleles for all these traits, and so will not require growth supplements. c. Diploid Aspergillus spores will produce green colonies with haploid or diploid sectors of white and yellow (green are also produced but are not detectable). Spore size indicates whether the sector is haploid or diploid. d. Haploidization (mitosis without chromosome replication) produces haploid segregants (haploid progeny nuclei). Haploid white sectors provide an example: i. Half the haploid white sectors have the genotype wad+propaba+y+bi and the other half w+adpro+pabaybi+. Except for the w allele, these are reciprocals. ii. This indicates that w is on one chromosome, and the five others are on a different chromosome, although gene order cannot be determined from haploid segregants. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 23 e. Diploid (mitotic) segregants are rare, and so are usually single crossovers. All genes distal to the crossover point on the chromosome arm are homozygous, allowing the recessive phenotypes to be seen. Diploid yellow sectors are an example (Figure 16.9). i. Mitotic crossover between pro and paba will produce a segregant homozygous for the y allele (y+/y+ twin spot is green and so not detected). The yellow segregant will require PABA acid to grow (Figure 16.10). ii. Mitotic crossover between paba and y will also produce a yellow sector, but these will not require PABA for growth. iii. Gene order, therefore, may be determined by mitotic crossover experiments with markers distal to the chromosome. Counting diploid segregants allows computation of map distance between genes (similar to meiotic recombination studies). 5. Genetic recombination without regular alternation of meiosis and fertilization are parasexual systems. In fungi such as Aspergillus, the parasexual cycle is: a. Formation of heterokaryon. b. Fusion in the heterokaryon of haploid nuclei of different genotypes to form heterozygous diploid nucleus. c. Mitotic crossing-over within the diploid nucleus. d. Haploidization of the diploid nucleus. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 24 Fig. 16.9 Possible mitotic crossing-over event between the pro and paba loci that can give rise to a diploid yellow sector in the green diploid Aspergillus strain of Figure 16.8 台大農藝系 遺傳學 601 20000 Chapter 13 slide 25 Fig. 16.10 Production of diploid yellow sector in a green diploid Aspergillus strain by mitotic crossing-over between the paba and y loci. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 26 Retinoblastoma, a Human Tumor That Can Be Caused by Mitotic Recombination 1. Retinoblastoma is the most common childhood eye cancer, occurring from birth to 4 years of age. Two types are known: a. The sporadic (nonhereditary) form occurs in an individual with no family history of the disease, and affects only one eye (unilateral). b. The hereditary form affects both eyes (bilateral) and usually occurs at an earlier age than sporadic. 2. A single gene (Rb) on chromosome 13q14 is involved. a. In hereditary retinoblastoma, tumor cells have mutations in both copies of this gene, while other cells in the same individual are heterozygous. The disease is caused by a second mutation that affects the normal RB allele. b. The second mutation is often identical to the one on the other chromosome, strong circumstantial evidence that the wild-type copy of the gene is somehow replaced by the inherited mutated allele. One possible explanation is mitotic recombination. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 27 Mapping Human Genes Mapping Human Genes by Recombination Analysis 1. Experimental crosses are not done in humans and so genetic mapping relies on pedigree analysis, but only a few multigenerational pedigrees are appropriate for analyzing autosomal linkage. Recombination analysis in humans is easier for X-linked genes. 2. In a theoretical example (Figure 16.11): a. A man with rare X-linked recessive alleles a and b marries a woman who expresses neither of these traits and is likely wild type (a+b+/a+b+). b. A daughter of this couple would be doubly heterozygous (a+b+/ab). She would produce recombinant gametes at a frequency related to the distance between the genes. c. If she pairs with a normal (a+b+/Y) man: i. All daughters will be wild type, due to inheriting a+b+ from their father. ii. Sons will express four phenotypic classes (parental a+b+ or ab, or recombinant a+b or ab+). d. If a large number of pedigrees are available, an estimate of genetic map distance may be obtained for these genes. 3. This approach has shown that the distance between the green weakness gene (g) responsible for a form of color blindness, and the hemophilia A gene (h) is eight map units. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 28 Fig. 16.11 Calculation of recombination frequency for the X-linked human genes by analyzing the male progenies of a woman doubly heterozygous for the two genes. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 29 lod Score Method for Analyzing Linkage of Human Genes 1. The lack of suitable human pedigrees showing segregation of defined linked traits makes mapping autosomes especially difficult, and so usually the lod (logarithm of odds) score method is used for statistical analysis of pedigree data. a. A lod score compares the expected distributions of traits if they are linked, and if they are not linked. b. The lod score is the log10 of the ratio of the two probabilities. The higher the lod score, the closer the two genes. 2. The map distance for linked markers is computed from the recombination frequency given by the highest lod score, by solving lod scores for a range of proposed map units. For the human genome, 1mu is approximately 1 megabase (Mb). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 30 High-Density Genetic Maps of the Human Genome 1. Human genetic mapping was revolutionized by: a. Discovery of many polymorphic DNA markers. b. Development of molecular tools to type the markers (determining molecular phenotypes). 2. Hundreds of markers may be typed in a given cross, and computer algorithms are used to determine linkage relationships. 3. A high-density human genetic map was completed in 1994. a. A consortium of laboratories worked on the same set of DNA samples (mapping panel), so their data could be combined. b. Localized 5,264 STR (short tandem repeat) markers to 2,335 chromosomal loci, with an average density of one marker per 599kb (Figure 16.12). 台大農藝系 遺傳學 601 20000 Chapter 13 slide 31 Fig. 16.12 A high density genetic map with 5,264 microsattelites localized to 2,335 chromosomal loci 台大農藝系 遺傳學 601 20000 Chapter 13 slide 32 Physical Mapping of Human Genes FISH (Fluorescent in situ Hybridization) Maps 1. Individual metaphase chromosomes are probed in situ with specific fluorescently labeled DNA sequences, identifying homologous sequences in the chromosome (Figure 16.13). 2. Different probes labeled with different fluorescent dyes may be used in the same experiment. Fluorescence microscopy provides data for computer imaging analysis to determine the binding site(s) for each probe. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 33 Fig. 16.13 An example of the results of fluorescent in situ hybridization (FISH) in which fluorescently tagged DNA probes were hybridized to human metaphase chromosome 台大農藝系 遺傳學 601 20000 Chapter 13 slide 34 Radiation Hybrid Maps 1. A radiation hybrid (RH) is a rodent cell line carrying a small genomic DNA molecule from another organism (e.g., a human). In this technique (Figure 16.14): a. Exposure to X-rays breaks the DNA in human cells. The fragments become smaller with more X-ray exposure, and fragment length determines the map resolution. b. Irradiation kills the human cells, which are then fused with rodent cells, rescuing chromosomal fragments that are typically a few Mb in length. c. Human DNA in the RH is analyzed for gene and/or DNA markers. The closer two markers are to each other on the chromosome, the more likely they are to be found together in an RH. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 35 Fig. 16.14 Making the radiation hybrid. 台大農藝系 遺傳學 601 20000 Chapter 13 slide 36