* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download UCLA Bioinformatics - Cal State LA
Signal transduction wikipedia , lookup
Phosphorylation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein design wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein domain wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein folding wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
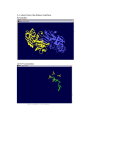
Exploring the Biology of Disulfide-Rich Hyperthermophiles through Protein Phylogenetic Profiles Navapoln Ramakul1, Morgan Beeby12, and Todd O. Yeates123 1Department of Chemistry and Biochemistry, 2Department of Energy Center for Genomics and Proteomics, and 3Molecular Biology Institute, University of California, Los Angeles, CA 90095-1569 Southern California Bioinformatics Summer Institute UCLA Bioinformatics: Yeates Lab • Goals: determine and analyze the threedimensional structures of proteins. • Research: focus on protein structure & function, protein sequence & evolution, and protein assembly & design. • Methods: crystal structure determination through theoretical and computational methods. General Overview • Genomic Databases: create opportunities for new kinds of computational analyses and novel discoveries. • Advantage: special comparative studies using multiple genomes to compare sequence vs. structure. • Present Research: investigate the surprising revelation about disulfide bonds in certain microbes from comparative studies. Protein Disulfide Bonds • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Previously believed to be prominent only outside the cell. Inside the cell • Disulfides only rarely found. • Disulfides are transient or functionally important, rather than stabilizing. • Summary Outside the cell • Abundant. Recent Studies • Intro • Comput. Methods • Results & Significance • Unexpected disulfides in an intracellular protein. • Crystal structure of adenylosuccinate lyase (ASL) from the P. aerophilum surprisingly shown a protein chain stabilized by three disulfide bonds (Toth et al., JMB (2000) 301, 433-450.). • Applications & Future Directions Disulfide bond • Summary Disulfide bond Toth et al., JMB (2000) 301, 433-450 Evidence for Abundant S-S bonds in P. aerophilum • Intro • Comput. Methods • Results & Significance Proteins with Even # of Cysteines • Applications & Future Directions • Summary Mallick, Boutz, Eisenberg, and Yeates (2002). PNAS 99, 9679-9684 Disulfide Abundance in Various Genomes Genome S) Archaeal branch f (S- Pyrobaculum aerophilum 0.44 Aeropyrum pernix 0.40 Pyrococcus abyssi 0.31 Pyrococcus horikoshii 0.28 Aquifex aeolicus 0.17 Genome f (S-S) Pyro. aerophilum 0.44 104°C Aero. pernix 0.40 100°C Pyro. abyssi 0.31 102°C Pyro. horikoshii 0.28 102°C Aqui. aeolicus 0.17 93°C Meth. thermo 0.15 90°C Methanobacterium thermo 0.15 Thermotoga maritima 0.13 Methanococcus jannasc 0.13 Archaeoglobus fulgidus 0.11 Mycoplasma genitalium 0.06 Synechocystis PCC6803 0.08 Ureaplasma urealyticum 0.07 Neisseria meningitidis 0.06 Mycobacterium tubercu 0.07 Rickettsia prowazekii 0.06 Haemophilus influenzae 0.05 Escherichia coli 0.05 Treponema pallidum 0.03 Helicobacter pylori 0.03 Bacillus subtilis 0.01 90°C 86°C 92°C Blue = archaea = thermophile Eubacterial branch Mallick, Boutz, Eisenberg, and Yeates (2002). PNAS 99, 9679-9684 Exploring disulfide-rich hyperthermophiles • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Summary • Find the sequences of glutaredoxin-like protein in different organisms. • Investigate the glutaredoxin-like protein in those disulfide-rich hyperthermophiles. • Goals: differences between glutaredoxin-like protein in hyperthermophiles and glutaredoxin in organisms. Why glutaredoxin-like protein? • Only present among hyperthermophiles. • Operates in thiol-disulfide reaction via CXXC motif which either form a disulfide (oxidized form) or a dithiol (reduced form). • Requires for many functions including electron and proton transport to essential enzymes like ribonucleotide reductase. • Involves in formation of disulfide bonds in protein folding. 90o Prototypical fold: E.coli thioredoxin (2TRX.pdb) Methods • Intro • Comput. Methods • The sequences used in this study were obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). • Results & Significance • Obtain the control sequence of glutaredoxin (E. coli) to find glutaredoxin-like protein. • Applications & Future Directions • Search for the glutaredoxin-like protein sequences of hyperthermophilic archaea. • Summary • Using Sequence-Structure Mapping to identify potential disulfide bonds. • Compare and analyze using multiple sequences alignment program, such as ClustalW, T-Coffee, or MSA. Results • ClustalW multiple sequences alignment of these glutaredoxin-like proteins shows two CXXC motifs Green = P. aerophilum Black = Hyperthermophilic archeae Blue = Bacteria = CXXC motif Pyrobaculum_aerophilum Aeropyrum_pernix Pyrococcus_abyssi Pyrococcus_horikoshii Aquifex_aeolicus Thermotoga_maritima Glutaredoxin_1 Glutaredoxin_3 Bacillus_subtilis Prim.cons. 10 20 30 40 50 60 70 80 90 100 110 120 130 140 | | | | | | | | | | | | | | MAVPIGGPEEVPHIEVDEETKEIIKEMLSQMENPVNINFFTSPNCAGRETNWCIPTEELLDLLVQLAPQ-----GKLI-VNKYNAEKDVEAFKKFGVEPQRVPVIYFGE--GF-IRYLGAPMGEEVRAFIETVVRLSTGK MAR-------YYVLDLSEDFRRELRETLAEMVNPVEVHVFLSK—SGCET--CEDTLRLMKLFEEESPTR--NGGKLLKLNVYYRESDSDKFSEFKVE--RVPTVAFLG--GE-VRWTGIPAGEEIRALVEVIMRLSEDE ----------MGLISEEDKRI-IKEEFFSKMVNPVKLIVFIG----KEHCQYCDQLKQLVQELSELT------DKLSYEIVDFDTPEGKELAEKYRIDRAP-ATTITQDGKDFGVRYFGIPAGHEFAAFLEDIVDVSRAE ----------MGLISEEDKRI-IKEEFFSKMVNPVKLIVFIG----KEHCQYCDQLKQLVQELSELT------DKLSYEIVDFDTPEGKELAEKYRIDRAP-ATTITQDGKDFGVRYFGIPAGHEFAAFLEDIVDVSKGD ------------MLLNLDVRMQLKELAQKEFKEPVSIKLFS----QAIGCESCQTAEELLKETVEVIGEAVGQDKIKLDIYSPFT--HKEETEKYGVDRVP-TIVIEGD-KDYGIRYIGLPAGLEFTTLINGIFHVSQRK ----------MGILSDKDIAY-LKDLFGKELKRKVKIVFFKTE--DKTRCQYCEITEQVLEELVSVD------PKLELEIHDFDS--DKEAVEKYQVEMVPATILLPEDGKDYGIRFYGVPSGHEFGTLIQDIITVSEGK ----------------------------------MQTVIFG-----RSGCPYCVRAKDLAEKLSNER--------------------DDFQYQYVDIRA---EGITKED----LQQKAGKPV-ETVPQIFVDQQHIGGYT ---------------------------------MANVEIYT-----KETCPYCHRAKALLS-----S--------------------KGVSFQELPIDG---NAAKREE----MIKRSGR---TTVPQIFIDAQHIGGCD ----------------------------------MRLIKLE-----QPNCNPCKMVSNYLEQVN------------------------IQ-FETVDVTQ---EPEVAAR-----FGVMGVP----VTILLSDQGEEVNRS * . . : * . :. MA2PIGGPEEMGL2SEEDKRI3IKEEF2SEMVNPVK2IVF223NC4KE3CQYC222KQLLEEL2EL2P32VG2DKL2LEI2DFDT2EDKE2FEKY3VDR2PV4T3I2EDGKDFGIRYFGIPAG2E24AL2EDIVHVS2GK Two CXXC motifs Pyrobaculum_aerophilum Aeropyrum_pernix Pyrococcus_abyssi Pyrococcus_horikoshii Aquifex_aeolicus Thermotoga_maritima Glutaredoxin_1 Glutaredoxin_3 Bacillus_subtilis Prim.cons. 150 160 170 180 190 200 210 220 230 240 250 260 270 | | | | | | | | | | | | | TGLRQKTRAE-LSTLAQGAPKRVYILTVVTPSCPYCPYAVLMANMFAYES--KGK--VVSVVVEAYENPDIADMYGVTGVPTVILQAEDAAVGDVEFVGVPPEHELLA-------RVKNHMG--LS------------SGLEDATKEA-LKSLKG----RVHIETIITPSCPYCPYAVLLAHMFAYEAWKQGNPVILSEAVEAYENPDIADKYGVMSVPSIAIN------GYLVFVGVPYEEDFLD-------YVKSAAEGRLTVKGPIRAGEAEEL TDLMAESKEE-VAKIDKN----VRILVFVTPTCPYCPLAVRMAHKFAIENTKAGKGKILGDMVEAIEYPEWADQYNVMAVPKIVIQVDGE--DKVQFEGAYPEKMFLE-------KLLAALS----------------TDLMQDSKEE-VSKIDKD----VRILIFVTPTCPYCPLAVRMAHKFAIENTKAGKGKILGDMVEAIEYPEWADQYNVMAVPKIVIQVNGE--DKVQFEGAYPEKMFLE-------KLLSALS----------------PQLSEKTLEL-LQVVDIP----IEIWVFVTTSCGYCPSAAVMAWDFALAN-----DYITSKVIDASENQDLAEQFQVVGVPKIVINKG-----VAEFVGAQPENAFLGYIMAVYEKLKREKEQA--------------PQLSEESIQK-LQSLEEP----IRISVFVTPTCPYCPRAVLMAHNMAMAS-----DKIIGEMIEANEYWELSEKFGVSSVPHIVVNRDP----SKFFVGAYPEKEFIN------EVLRLAKG----------------DFAAWVKEN--LDA----------------------------------------------------------------------------------------------------------------------------DLYALDARGG-LDPLLK-------------------------------------------------------------------------------------------------------------------------VGFKPNELDELLKELR--------------------------------------------------------------------------------------------------------------------------: TGL3232KEELL42LDKPAPKRVRILVFVTP2CPYCP2AVLMAH2FA2ENTKAGK2KIL22MVEA2E2P23ADQYGVM3VPKIVI2VDGEAV2KV2FVGAYPEK2FLEYIMAVYEKLKSA2322L2VKGPIRAGEAEEL • Glutaredoxin-like protein has more than 85 amino acids. Results • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Most organisms have 1 CXXC motif in glutaredoxin. • Glutaredoxin-like protein has two redox-active CXXC motifs per polypeptide. • Exception: P. aerophilum has only 1 CXXC motif. P. furiosus: P. horikoshii: • Summary CXXC motifs 1A8L.pdb, Nat. Struct. Biol. (1998), 5 (7) 602-611 1J08.pdb, unpublished Limitation • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Summary • Only 25 genomes. • Some glutaredoxin-like proteins have not yet been sequenced. Applications and Further Studies: • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Summary • How disulfide bonds involve in protein folding? • To identify disulfide-bonded protein-protein interactions and networks. • To investigate the stability mechanisms by disulfide bonds. Summary • Intro • Comput. Methods • Results & Significance • Applications & Future Directions • Summary • Most of the hyperthermophiles have 2 CXXC motifs in order to have abundant disulfide bonds. • The abundance of disulfide bonds appear to play a key role in stabilizing protein at high temperature. • Intracellular disulfide bond is a characteristic of all archaea or an adaptation to high temperature. • This study illustrates the power of integrating genomic data with protein structure and function to illuminate the chemistry and biology of unusual organisms. Acknowledgements Yeates Lab Dr. Todd Yeates Morgan Beeby Everyone else at Yeates lab CalState LA Research mentors SoCalBSI Program SoCalBSI interns Support National Science Foundation (NSF) National Institutes of Health (NIH). UCLA-DOE Center for Genomics and Proteomics