* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Nucleic Acids - Farmasi Unand
Molecular cloning wikipedia , lookup
Polyadenylation wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Non-coding DNA wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Protein adsorption wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Proteolysis wikipedia , lookup
Bottromycin wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Messenger RNA wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression wikipedia , lookup
Biochemistry wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Expanded genetic code wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genetic code wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
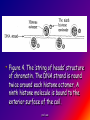
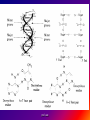
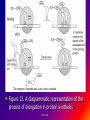
Reference: Gareth Thomas Week 11, 12,13 prof. aza 1. Introduction • The nucleic acids are the compounds that are responsible for the storage and transmission of the genetic information that controls the growth, function and reproduction of all types of cells. prof. aza • They are classified into two general types: the deoxyribonucleic acids (DNA), whose structures contain the sugar residue β-D-deoxyribose; and the ribonucleic acids (RNA), whose structures contain the sugar residue β -D-ribose (Figure 10.1). prof. aza Figure 1. The structures of β -D-deoxyribose and β -D-ribose. prof. aza nucleotide consists of a purine or pyrimidine base • Both types of nucleic acids are polymers based on a repeating structural unit known as a nucleotide (Figure 10.2). These nucleotides form long single-chain polymer molecules in both DNA and RNA. • Each nucleotide consists of a purine or pyrimidine base bonded to the 1’ carbon atom of a sugar residue by a β -Nglycosidic link (Figure 10.3). prof. aza • These base-sugar subunits, which are known as nucleosides, are linked through the 3’ and 5’ carbons of their sugar residues by phosphate units to form the polymer chain. prof. aza Figure 2. The general structures of (a) nucleotides and (b) a schematic representation of a section of a nucleic acid chain. prof. aza Figure 3. Examples of the structures of some of the nucleosides found in RNA. The β -Nglycosidic link is shaded The corresponding nucleosides in DNA are based on deoxyribose and use the same name but with the prefix deoxy. prof. aza 2. Deoxyribonucleic Acids (DNA) • DNA occurs in the nuclei of cells in the form of a very compact DNA protein complex called chromatin. The protein in chromatin consists mainly of histones, a family of relatively small positively charged proteins. prof. aza • The DNA is coiled twice around as octomer of histone molecules with a ninth histone molecule attached to the exterior of these mini coils to form a structure like a row of heads spaced along a string (Figure 10.4). prof. aza • This ‘string of beads’ is coiled and twisted into compact structures known as miniband units, which form the basis of the structures of chromosomes. Chromosomes are the structures that form duplicates during cell division in order to transfer the genetic information of the old cell to the two new cells. prof. aza 2.1 Structure DNA molecules are large with relative molecular masses up to one trillion. The principal bases found in their structures are adenine (A), thymine (T), guanine (G) and cytosine (C), although derivatives of these bases are found in some DNA molecules (Figure 10.5). Those bases with an oxygen function have been shown to exist in their keto form. prof. aza • Figure 4. The ‘string of heads’ structure of chromatin. The DNA strand is round twice around each histone octomer. A ninth histone molecule is bound to the exterior surface of the coil. prof. aza DNA-binding proteins Interaction of DNA with histones (shown in white, top). These proteins' basic amino acids (below left, blue) bind to the acidic phosphate groups on DNA (below right, red). prof. aza Figure 5. The purine and pyrimidine bases found in DNA. The numbering is the same or each type of ring system. prof. aza Structures of the four bases found in DNA and the nucleotide adenosine monophosphate. Adenine Guanine Adenosine monophosphate Thymine Cytosine prof. aza • Chargaff showed that the molar ratios of adenine to thymine and guanine to cytosine are always approximately 1: 1 in any DNA structure although the ratio of adenine to guanine varies according to the species from which the DNA is obtained. prof. aza • This and other experimental observations lead Crick and Watson in 1953 to propose that the threedimensional structure of DNA consisted of two single molecule polymer chains held together in the form of a double helix by hydrogen bonding between the same pairs of bases, namely: prof. aza • the adenine- thymine and cytosine- guanine base pairs (Figure 10.6). These pairs of bases, which are referred to as complementary base pairs, form the internal structure of the helix. prof. aza • They are hydrogen bonded in such a manner that their flat structures lie parallel to one another across the inside of the helix. The two polymer chains forming the helix are aligned in opposite directions. In other words, at the ends of the structure one chain has a free 3’OH group and the other chain has a free 5’-OH group. prof. aza • X-ray diffraction studies have since confirmed that this is the basic three dimensional shape of the polymer chains of the β -DNA, the natural form of DNA. • This form of DNA has about ten bases per turn of the helix. prof. aza • Its outer surface has two grooves known as the minor and major grooves, respectively, which act as the binding sites for many ligands. Two other forms of DNA, the A and Z forms, have also been identified but it is not certain if these forms occur naturally in living cells. prof. aza • Electron microscopy has shown that the double helical chain of DNA is folded, twisted and coiled into quite compact shapes. A number of DNA structures are cyclic and these compounds are also coiled and twisted into specific shapes. These shapes are referred to as supercoils, supertwists and superhelices as appropriate. prof. aza prof. aza The two strands of DNA are held together by hydrogen bonds between bases. The sugars in the backbone are shown in light blue. prof. aza • Figure 10.6. The double helical structure of B-DNA. Interchanging of either the bases of a base pair and/or base pair with base pair does not affect the geometry of this structure. Reproduced from G. Thomas. (Chemistry for Pharmacy and the Life Sciences including Pharmacology and Biomedical Science, I996, by per mission of Prentice Hall, a Pearson Education Company. prof. aza 3. The General Functions of DNA The DNA found in the nuclei of cells has three functions: (i) to act as a repository for the genetic information required by a cell to reproduce that cell: (ii) to reproduce itself in order to maintain the genetic pool when cells divide; (iii) to supply the information that the cell requires to manufacture specific proteins. prof. aza • Genetic information is stored in a form known as genes by the DNA found in the nucleus of a cell (see section 10.4). • The duplication of DNA is known as replication. It results in the formation of two identical DNA molecules that carry the same genetic information from the original cell prof. aza • to the two new cells that are formed when a cell divides (see section 10.5). • The function of DNA in protein synthesis is to act as a template for the production of the various RNA molecules necessary to produce a specific protein (see section 6) prof. aza Figure 10.7. A schematic representation of the gene for the β -subunit of haemoglobin. prof. aza 4. Genes • Each species has its own internal and external characteristics. These characteristics are determined by the information stored and supplied by the DNA in the nuclei of its cells. • This information is carried in the form of a code based on the consecutive sequences of bases found in sections of the DNA structure (see section 5). prof. aza • This code controls the production of the peptides and proteins required by the body. • The sequence of bases that act as the code for the production of one specific peptide or protein molecule is known as a gene. prof. aza Changing the sequence of the bases effect on the external or internal characteristics of an individual • Genes can normally contain from several hundred to 2000 bases. Changing the sequence of the bases in a gene by adding, subtracting or changing one or more bases may cause a change in the structure of that protein with a subsequent knock-on effect on the external or internal characteristics of an individual. prof. aza • For example, an individual may have brown instead of blue eyes or their insulin production may be inhibited, which could result in that individual suffering from diabetes. • A number of medical conditions have been attributed to either the absence of a gene or the presence of a degenerate or faulty gene in which one or more of the bases in the sequence have been changed. prof. aza • In simple organisms, such as bacteria, genetic information is usually stored in a continous sequence of DNA bases. • However, in higher organisms the bases forming a particular gene may occur in a number of separate sections known as exons, separated by sections of DNA that do not appear to be a code for any process. prof. aza • These non-coding sections are referred to as introns. For example, the gene responsible for the β-subunit of haemoglobin consists of 990 bases. These bases occur as three exons separated by two introns (Figure 10.7). prof. aza • The complete set of genes that contain all the hereditary information of a particular species is called a genome. • The Human Genome Project. initiated in 1990, sets out to identify all the genes that occur in human chromosomes and also the sequence of bases in these genes. prof. aza • This will create an index that can be used to locate the genes responsible for particular medical conditions. For example, the gene in region q31 of chromosome 7 is responsible for the protein that controls the flow of chloride ions through the membranes in the lungs. prof. aza • The changing of about three bases in exon number 10 gives a degenerate gene that is known to be responsible for causing cystic fibrosis in a large number of cases. prof. aza Figure 10.8. A schematic representation of the replication of DNA. The arrows show the direction of growth of the leading and lagging strands. Reproduced from G. Thomas, Chemistry for Pharmacy and the Life Sciences including Pharmacology and Biomedical Science, 1996, by permission of Prentice Hall, a Pearson Education Company. prof. aza • Figure 10.8. A schematic representation of the replication of DNA. The arrows show the direction of growth of the leading and lagging strands. • Reproduced from G. Thomas, Chemistry for Pharmacy and the Life Sciences including Pharmacology and Biomedical Science, 1996, by permission of Prentice Hall, a Pearson Education Company. prof. aza DNA replication. The double helix (blue) is unwound by a helicase. Next, DNA polymerase III (green) produces the leading strand copy (red). A DNA polymerase I molecule (green) binds to the lagging strand. This enzyme makes discontinuous segments (called Okazaki fragments) before DNA ligase (violet) joins them together. prof. aza 5. Replication • Replication is believed to start with the unwinding of a section of the double helix (Figure 10.8). • Unwinding may start at the end or more commonly in a central section of the DNA helix. It is initiated by the binding of the DNA to specific receptor proteins that have been activated by the appropriate first messenger (see section 8.4). prof. aza • The separated strands of the DNA act as templates for the formation of a new daughter strand. • Individual nucleotides, which are synthesised in the cell by a complex route, bind by hydrogen bonding between the bases to the complementary parent nucleotides. prof. aza • This hydrogen bonding is specific: only the complementary base pairs can hydrogen bond. • In other words, the hydrogen bonding can only be between either thymine and adenine or cytosine and guanine. prof. aza • This means that the new daughter strand is an exact replica of the original DNA strand bound to the parent strand. • Consequently, replication will produce two identical DNA molecules. prof. aza • As the nucleotides hydrogen bond to the parent strand they are linked to the adjacent nucleotide, which is already hydrogen bonded to the parent strand, by the action of enzymes known as DNA polymerases. • As the daughter strands grow, the DNA helix continues to unwind. • However, both daughter strands are formed at the same time in the 5’ to the 3’ direction. prof. aza • This means that the growth of the daughter strand that starts at the 3’ end of the parent strand can continue smoothly as the DNA helix continues to unwind. • This strand is known the leading strand. However, this smooth growth is riot possible for the daughter strand that started from the 5’ of the parent strand. prof. aza • This strand, known as the lagging strand, is formed in a series of sections, each of which still grows in the 5’ to 3’ direction. • These sections. which are known as Okazaki fragments after their discoverer, are joined together by the enzyme DNA ligase to form the second daughter strand. prof. aza • Replication, which starts at the end of a DNA helix, continues until the entire structure has been duplicated. • The same result is obtained when replication starts at the centre of a DNA helix. • In this case, unwinding continues in both directions until the complete molecule is duplicated. This latter situation is more common. prof. aza • DNA replication occurs when cell division is imminent. At the same time, new histones are synthesised. • This results in a thickening of the chromatin filaments into chromosomes (see section 2). These rod-like structures can be stained and are large enough to be seen under a microscope. prof. aza 6. Ribonucleic Acids (RNA) • Ribonucleic acids are found in both the nucleus and the cytoplasm. In the cytoplasm RNA is located mainly in small spherical organelles known as ribosome. These consist of about 65% RNA and 35% protein. • Ribonucleic acids are classified according to their general role in protein synthesis as: messenger RNA (mRNA): transfer RNA (tRNA): and ribosomal RNA (rRNA). prof. aza • Messenger RNA informs the ribosome as to what amino acids are required and their order in the protein, that is, they carry the genetic information necessary to produce a specific protein. • This type of RNA is synthesised as required and once its message has been delivered it is decomposed. prof. aza Figure 10.9. (a) The general structure ol a section of an RNA polymer chain. (b) The hydrogen bonding between uracil and adenine. Reproduced from G.’Thomas, Chemistry to Pharmacy and the Life Science including Pharmacology including Biomedical Science, 1996, by permission of Prentice Hall, a Pearson Education Company. prof. aza • Figure 10.9. (a) The general structure of a section of an RNA polymer chain. (b) The hydrogen bonding between uracil and adenine. Reproduced from G.’Thomas, Chemistry to Pharmacy and the Life Science including Pharmacology including Biomedical Science, 1996, by permission of Prentice Hall, a Pearson Education Company. prof. aza Figure 10.10. A schematic representation of a transcription process. Reproduced from G.’Thomas, Chemistry to Pharmacy and the Life Science including Pharmacology including Biomedical Science, 1996, by permission of Prentice Hall, a Pearson Education Company. prof. aza • The structures of RNA molecules consist of a single polymer chain of nucleotides with the same bases as DNA, with the exception of thymine, which is replaced by uracil ( Figure 9). • These chains often contain singlestranded loops separated by short sections of a distorted double helix (Figure 11). These structures are known as hairpin loops. prof. aza • All types of RNA are formed from DNA by a process known as transcription. It is thought that the DNA unwinds and the RNA molecule is formed in the 5’ to 3’ direction. • It proceeds smoothly. with the 3’ end of the new strand bonding to the 5’ end of the next nucleotide (Figure 10.10). prof. aza • This bonding is catalysed by enzymes known as RNA polymerases. • The sequence of bases in the new RNA strand is controlled by the sequence of bases in the parent DNA strand. • In this way DNA controls the genetic information being transcribed into the RNA molecule. • The strands of DNA also contain start and stop signals, which control the size of the RNA molecule produced. prof. aza • These signals are in the form of specific sequences of bases. • It is believed that the enzyme rho factor could be involved in the termination of the synthesis and the release of some RNA molecules from the parent DNA strand. However, in many cases there is no evidence that this enzyme is involved in the release of the RNA molecule. prof. aza • The RNA produced within the nucleus by transcription is known as heterogeneous nuclear RNA (hnRNA), premessenger RNA (premRNA) or primary transcript RNA ( ptRNA). prof. aza • Since the DNA gene from which it is produced contains both exons and introns, the hnRNA will also contain its genetic information in the form of a series of exons and introns complementary to those of its parent gene. prof. aza 7. Messenger RNA (mRNA) • mRNA carries the genetic message from the DNA in the nucleus to a ribosome. This message instructs the ribosome to synthesise a specific protein. • mRNA is believed to be produced in the nucleus from hnRNA by removal of the introns and the splicing together of the remaining exons into a continuous genetic message, prof. aza • the process being catalysed by specialised enzymes. The net result is a smaller mRNA molecule with a continuous sequence of bases that are complementary to the gene’s exons, this mRNA now leaves the nucleus and carries its message in the form of a code to a ribosome. prof. aza prof. aza all protein synthesis starts with methionine • The code carried by mRNA was broken in the 1960s by Nirenberg and other workers. These workers demonstrated that each naturally occurring amino acid had a DNA code that consisted of a sequence of three consecutive bases known as a codon and that an amino acid could have several different codons (Table 10.1), • In addition, three of the codons are stop signals which instruct the ribosome to stop protein synthesis. prof. aza • Furthermore, the codon that initiates the synthesis is always AUG, which is also the codon for methionine. Consequently, all protein synthesis starts with methionine. However, few completed proteins have a terminal methionine because this residue is normally removed before the peptide chain is complete. prof. aza • Moreover, methionine can still be incorporated in a peptide chain because there are two different tRNAs that transfer methionine to the ribosome (see section 8). prof. aza all living matter using the same genetic code for protein synthesis • One is specific for the transfer of the initial methionine whereas the other will only deliver methionine to the developing peptide chain, By convention, the three letters of codon triplets are normally written with their 5’ ends on the left and their 3’ ends on the right. prof. aza • The mRNA’s codon code is known as the genetic code, Its use is universal, all living matter using the same genetic code for protein synthesis. • This suggests that all living matter must have originated from the same source and is strong evidence for Darwin’s theory of evolution. prof. aza • Figure .11. The general structures of tRNA. (a) The two-dimensional cloverleaf representation showing some of the invariable nucleotides that occur in the same positions in most tRNA molecules and (b) the three- dimensional L shape (From CHEMISTRY, by Linus Pauling and Peter Pauling. Copyright © 1975 by Linus Pauling and Peter Paling. Used with permission of W. H. Freeman and Company) prof. aza 8. Transfer RNA (tRNA) • tRNAs are also believed to be formed in the nucleus from the hnRNA. • They are relatively small molecules that usually contain from 73 to 94 nucleotides in a single strand. Some of these nucleotides may contain derivatives of the principal bases, such as 2’-O-methylguanosine (0MG) and inosine (I). • The strand of tRNA is usually folded into a three-dimensional L shape. prof. aza • This structure, which consists of several loops, is held in this shape by hydrogen bonding between complementary base pairs in the stem sections of these loops and also by hydrogen bonding between bases in different loops. prof. aza Figure .11. The general structures of tRNA. (a) The two-dimensional cloverleaf representation showing some of the invariable nucleotides that occur in the same positions in most tRNA molecules and (b) the three- dimensional L shape (From CHEMISTRY, by Linus Pauling and Peter Pauling. Copyright © 1975 by Linus Pauling and Peter Paling. Used with permission of W. H. Freeman and Company) prof. aza • Figure .11. The general structures of tRNA. (a) The two-dimensional cloverleaf representation showing some of the invariable nucleotides that occur in the same positions in most tRNA molecules and (b) the three- dimensional L shape (From CHEMISTRY, by Linus Pauling and Peter Pauling. Copyright © 1975 by Linus Pauling and Peter Paling. Used with permission of W. H. Freeman and Company) prof. aza • This results in the formation of sections of double helical structures. • However, the structures of most tRNAs are represented in two dimensions as a cloverleaf (Figure 11). prof. aza • tRNA molecules carry amino acid residues from the cell’s amino acid pool to the mRNA attached to the ribosome. • The amino acid residue is attached through an ester linkage to ribosome residue at the 3’ terminal of the tRNA strand, which almost invariably has the sequence CCA. prof. aza • This sequence plus a fourth nucleotide projects beyond the double helix of the stem. Each type of amino acid can only be transported by its own specific tRNA molecule. In other words a tRNA that carries serine residues will not transport alanine residues. In other word, some amino acids can be carried by several different tRNA molecules prof. aza • The tRNA recognises the point on the mRNA where it has to deliver its amino acid through the use of a group of three bases known as an anticodon. • This anticodon is a sequence of three bases found on one of the loops of the tRNA (Figure 11). • The anticodon can only form base with the complementary codon in the mRNA. prof. aza prof. aza • Consequently, the tRNA will only hydrogen bond to the region of the mRNA that has the correct codon, which means its amino acid can only be delivered to a specific point on the mRNA. prof. aza • For example, a tRNA molecule with the anticodon CGA will only transport its alanine residue to a GCU codon on the mRNA. • Furthermore, this mechanism will also control the order in which amino acid residues are added to the growing protein. prof. aza 9. Ribosomal RNA (rRNA) • Ribosomes contain about 35% protein and 65% rRNA. • Their structures are complex and have not yet been fully elucidated. • However, they have been found to consist of two Sections that are referred to as the large and small subunits. • Each of these subunits contains protein and rRNA prof. aza • In Eschericia coli the small subunit has been shown to contain a 1542-nucleotide rRNA molecule whereas the large contains two rRNA molecules of 120 (Figure 12) and 2094 nucleotides, respectively. prof. aza • Experimental evidence suggests that rRNA molecules have structures that consist of a single strand of nucleotides whose sequence varies considerably from species to species. • The strand is folded and twisted to form a series of single-stranded loops separated by sections of double helix (Figure 12). prof. aza Figure 10.12. The proposed sequence of nucleotides in the 120-nucleotide subunit found in Escherichia coli ribosome showing the single-stranded loops and the double helical structures. (Reprinted, with permission, from the Annual Review of Biochemistry, volume 53 © I984 by Annual Reviews. www.Annual Reviews.org). prof. aza • The double helical segments are believed to be formed by hydrogen bonding between complementary base pairs. • The general pattern of loops and helixes is very similar between species even though the sequence of nucleotides are different prof. aza • However, little is known about the three-dimensional structures of rRNA molecules and their interactions with the proteins found in the ribosome. prof. aza 10. Protein Synthesis • Protein synthesis starts from the N-terminal of the protein. • It proceeds in the 5’ to 3’ direction along the mRNA and may be divided into four mayor stages. namely: activation: initiation: elongation: and termination. prof. aza Activation • Activation is the formation of the tRNA amino acid complex. • Initiation is the binding of the mRNA to the ribosome and the activation of the ribosome. Elongation is the formation of the protein. prof. aza • Termination is the ending of the protein synthesis and its release from the ribosome. • All these processes normally require the participation of protein catalysts, known as factors, as well as other proteins whose function is not always known. • GTP and sometimes ATP act as sources of energy for the processes. prof. aza 10.1 Activation It is believed that the amino acids from the cellular pool react with ATP to form an active amino acid-AMP complex. This complex reacts with the specific tRNA for the amino acid. the reaction being catalysed by a synthese that is specific for that amino acid. prof. aza • It is believed that the amino acids ( AA) from the cellular pool react with ATP to form an active amino acid-AMP complex (AA-AMP). • This complex reacts with the specific tRNA for the amino acid, the reaction being catalysed by a synthase that is specific for that amino acid. prof. aza Figure 13. A schematic representation of the initiation of protein synthesis. prof. aza 2. Initiation • The general mechanism of initiation is well documented but the liner details are still not known. • It is thought that it starts with the two subunits of the ribosome separating and the binding of the mRNA to the smaller subunit. • Protein synthesis then starts by the attachment of a methionine-tRNA complex to the mRNA so that it forms the N-terminal of the new protein. prof. aza • Methionine is always the first amino acid in all protein synthesis because its tRNA anticodon is also the signal for the ribosome system to start protein synthesis. • Because the anticodon for methionine tRNA is UAC, this synthesis will start at the AUG codon of the mRNA. prof. aza • This codon is usually found within the first 30 nucleotides of the mRNA. • However, few proteins have an Nterminal methionine because once protein synthesis has started the methionine is usually removed by hydrolysis. prof. aza • As soon as the methionine-tRNA has bound to the mRNA the larger ribosomal subunit is believed to bind to the smaller subunit so that the mRNA is sandwiched between the two subunits (Figure 13). • This large subunit is believed to have three binding sites called the P (peptidyl), A (acceptor) and E (exit) sites. prof. aza • It attaches itself to the smaller subunit so that its P site is aligned with the methionine- tRNA complex bound to the mRNA. • This P site is where the growing protein will be bound to the ribosome. prof. aza • The A site, which is thought to be adjacent to the P site, is where the next amino acid-tRNA complex binds to the ribosome so that its amino acid can be attached to the peptide chain. • The E site is where the discharged tRNA is transiently bound before it leaves the ribosome. prof. aza • This large subunit is believed to have three binding sites called the P (peptidyl), A (acceptor) and E (exit) sites. • It attaches itself to the smaller subunit so that its P site is aligned with the methionine-tRNA complex bound to the mRNA. prof. aza • This P site is where the growing protein will be bound to the ribosome. prof. aza • The A site, which is thought to be adjacent to the P site, is where the next amino acid-tRNA complex binds to the ribosome so that its amino acid can be attached to the peptide chain. • The E site is where the discharged tRNA is transiently bound before it leaves the ribosome prof. aza 3. Elongation • Elongation is the formation of the peptide chain of the protein by a stepwise repetitive process. • A great deal is known about the nature of this process but its exact mechanism is still not fully understood. prof. aza • The process of elongation is best explained by the use of a hypothetical example. • Suppose that the sequence of codons, including the start codon, is AUGUUGGCUGGA.. etc prof. aza • The elongation process starts with the methionine-tRNA bound to the AUG codon of the mRNA (Figure 14). • Because the second codon is UUG the second amino acid in the polypetide chain will be leucine. prof. aza • This amino acid is transported by a tRNA molecule with the anticodon AAC because this is the only anticodon that matches the UUG codon on the mRNA strand. • The leucine- tRNA complex ‘docks’ on the UUG codon of the mRNA and binds to the A site. prof. aza • This docking and binding is believed to involve ribosome proteins, referred to as elongation factors, and energy supplied by the hydrolysis of guanosine triphosphate (GTP) to guanosine diphosphate (GDP). • Once the leucine-tRNA has occupied the A site the methionin is linked to the leucine by means of a peptide link whose carbonyl group originates from the methionine. prof. aza • This reaction is catalysed by the appropriate transferase. • It leaves the tRNA on the P site empty and produces an (NH2)-MetLeu-tRNA complex at the A site. prof. aza • The empty tRNA is discharged through the E site and at the same time the complete ribosome moves along the mRNA in the 5’ to 3’ direction so that the dipeptidetRNA complex moves from the A site to the P site. prof. aza • This process is known as translocation. • It is poorly understood but it leaves the A site empty and able to receive the next amino acid tRNA complex. • The whole process is then repeated in order to add the next amino acid residue to peptide chain. prof. aza • Because the next mRNA codon in our hypothetical example is (GCU) this amino acid will be alanine (see Table 10. l). Subsequent amino acids are added in a similar way, the sequence of amino acid residues in the chain being control led by the order of the codons in the mRNA. prof. aza • It is poorly understood but it leaves the A site empty and able to receive the next amino acid tRNA complex. The whole process is then repeated in order to add the next amino acid residue to peptide chain . prof. aza • Figure 13. A diagrammatic representation of the process of elongation in protein synthesis prof. aza 10.4 Termination • The elongation process continues until a stop codon is reached. • This codon cannot accept an amino acid-tRNA complex and so the synthesis stops. • At this point the peptide-tRN chain occupies a P site and the A site is empty. prof. aza • The stop codon of the mRNA is recognised by proteins know as release factors, which promote the release of the protein from the ribosome. prof. aza • The mechanism by which this happens is not fully understood but they are believed convert the transferase responsible for peptide synthesis into a hydrolase, which catalyses hydrolysis of the ester group linking the polypeptide to its tRNA. prof. aza • Once released, the protein is folded into its characteristic shape. often under the direction of molecular chaperone protein. prof. aza 11 Protein Synthesis in Prokaryotic and Eukaryotic Cells • The general sequence of events protein synthesis is similar for both eukaryotic and pro prokaryotic cells. • In both cases the hydrolysis of GDP to GDP is the source of energy for many of the processes involved. • However, the structures of prokaryotic and eukaryotic ribosomes are different prof. aza • For example, the ribosomes of prokaryotic cells of bacteria are made up of 50S (see Apendix 3) and 30S rRNA subunits whereas the ribosomes of mammalian eukaryotic cells consist ,of 60S and 40S rRNA subunits. prof. aza • The differences between the ribosomes of prokaryotic and eukaryotic ribosomes are the basis of the selective action of some antibiotics . prof. aza 11.1. Prokaryotic CeLLs • The first step in protein synthesis is the correct alignment of mRNA on the small subunit of the ribosome. • In prokaryotic cells this alignment is believed to be due to binding by base pairing between bases at the 3’ end of the rRNA of the ribosome and bases at the 5’ end of the mRNA. prof. aza • This ensures the correct alignment of the AUG anticodon of the mRNA with the P site of the ribosome. • The mRNA sequence of bases responsible for this binding occurs as part of the upstream (5’ terminal end) section of the strand before the start codon. prof. aza • This sequence is often known as the Shine-Dalgarno sequence after its discovers. Shine-Dalgarno sequences vary in length and base sequence (Figure 10.15). • The initiating tRNA in prokaryotic cells is a specific methionine-tRNA known as tRNAfMet ,which is able to read the start codon AUG but not when it is part of the elongation sequence. tRNAfMet is unique in that the methionine it carries is usually in the form of its N-formyl derivative. prof. aza Figure 10.15. Examples of Shine-Dalgarno sequences (bold larger type) of mRNA recognised by Escherichia coli ribosomes. These sequences lie about 10 nucleotides upstream of the AUG start codon for the specified protein. prof. aza When AUG is part of the elongation sequence methionine is added to the growing protein by a different transfer RNA known as tRNAmMet, which also has the anticodon UAC. prof. aza • However, tRNAmMet cannot initiate protein synthesis. Elongation follows the general mechanism for protein synthesis (see section 10.9). • It requires a group of proteins known as elongation factors and energy supplied by the hydrolysis of GTP to GDP. Termination normally involves three release factors. prof. aza • Experimental work has shown that an mRNA strand actively synthesizing proteins still have several ribosomes attached to it at different places along its length. These multiple ribosome structures are referred to as polyribosomes or polysomes. prof. aza • The polysomes of prokaryotic cells can contain up to 10 ribosomes at any one lime. Each of these ribosomes will be simultaneously producing the same polypeptide or protein; prof. aza • the further the ribosome has moved along the mRNA, the longer the polypeptide chain. The process resembles the assembly line in a factory. Each mRNA strand can in its lifetime produce up to 300 protein molecules. prof. aza 10 amino acid residues are added.. • In prokaryotic but not eukaryotic cells (see section 4.1), ribosomes are found in association with DNA. • This is believed to he due to the ribosome binding to the mRNA as it is produced by transcription from the DNA. prof. aza • Furthermore, these ribosomes have been shown to start producing the polypeptide chain of their designated protein before transcription is complete. prof. aza • This means that in bacteria protein synthesis can be very rapid and in some cases faster than transcription. It has been reported that in some bacteria an average of 10 amino acid residues are added to the peptide chain ever second prof. aza 11.2. Eukaryotic Cells • The initiation of protein synthesis in eukaryotic cells follows a different route from that found in prokaryotic cells although it still uses a methionine-tRNA to start the process. prof. aza • Eukaryotic mRNAs has no Shine- Dalgarno sequences but are characterised by a 7-methyl GTP unit at the 5’ end of the mRNA strand and a polyadenosine nucleotide tail at the 3’ end of the strand (Figure 10.16). prof. aza prof. aza • In eukaryotic cells, the initiating tRNA is a unique form of the activated methionine- tRNA (tRNAi Met). However, unlike in the case of prokaryotic cells, the methionine residue it carries is not formylated. prof. aza • The initiating process is started by this tRNAi Met binding to the 40S subunit of the ribosome to form the so-called preinitiation complex, the process requiring the formation of a complex between tRNAi Met , various eukaryotic initiation factors (elFs) and GTP. prof. aza • At this point the mRNA binds to the 40S preinitiation complex. This binding process is believed to involve a number of eukaryotic initiation factors and energy supplied by the conversions of GTP to GDP and ATP to ADP. Once the mRNA has bound to the preinitiation complex the 60S subunit recombines with the 40S prof. aza • Once the mRNA has bound to the preinitiation complex the 60S subunit recombines with the 40S unit to form the initiation complex (Figure 10.17). prof. aza • The initiating process is started by this tRNAi Met binding to the 40S subunit of the ribosome to form the so-called preinitiation complex, the process requiring the formation of a complex between tRNAi Met , various eukaryotic initiation factors (elFs) and GTP. prof. aza • The absence of the Shine-Dalgarno sequence means that an alternative mechanism must he available to align the first AUG codon of the mRNA with the P site of the ribosome. This mechanism is believed to direct the preinitiation complex to the first AUG codon of the mRNA. prof. aza • Elongation in eukaryotic ribosomes follows the general mechanism for protein synthesis (see section 10.10.3) but involves different factors and proteins from those utilised by prokaryotic ribosomes. Termination only requires one release factor, unlike in prokaryotic ribosomes-where three release factors are usually required. prof. aza • Elongation in eukaryotic ribosomes follows the general mechanism for protein synthesis (see section 10.10.3) but involves different factors and proteins from those utilised by prokaryotic ribosomes. • Termination only requires one release factor, unlike in prokaryotic ribosomeswhere three release factors are usually required. prof. aza • Termination only requires one release factor, unlike in prokaryotic ribosomes-where three release factors are usually required. prof. aza Figure 10.17. An outline of the formation of the protein synthesis initiation complex by the ribosomes of eukaryotic cells. prof. aza