* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Silencer (genetics) wikipedia , lookup
Molecular evolution wikipedia , lookup
Magnesium transporter wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression wikipedia , lookup
Peptide synthesis wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Genetic code wikipedia , lookup
List of types of proteins wikipedia , lookup
Western blot wikipedia , lookup
Bottromycin wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Protein adsorption wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Rosetta@home wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein design wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Drug design wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Structural alignment wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Self-assembling peptide wikipedia , lookup
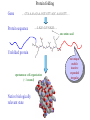
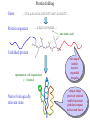
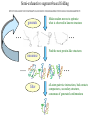
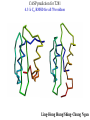
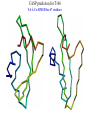
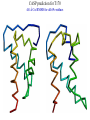
Computational engineering of bionanostructures Ram Samudrala University of Washington How can we analyse, design, & engineer peptides capable of specific binding properties and activities? A comprehensive computational approach • Sequence-based informatics - analyse sequence patterns responsible for binding specificity within experimentally characterised binders by creating specialised similarity matrices • Structure-based informatics - analyse structural patterns within experimental characterised binders by performing de novo simulations both in the presence and absence of substrate • Computational design - use de novo protocol to predict structures of the best candidate peptides or peptide assemblies, with validation by further experiment Sequence-based informatics • Create specialised similarity matrices by optimising the alignment scores such that strong, moderate, and weak binders for a given inorganic substrate cluster together – determines sequences patterns: Ersin Emre Oren (Sarikaya group) Protein folding Gene …-CTA-AAA-GAA-GGT-GTT-AGC-AAG-GTT-… Protein sequence …-L-K-E-G-V-S-K-D-… one amino acid Unfolded protein spontaneous self-organisation (~1 second) Native biologically relevant state not unique mobile inactive expanded irregular Protein folding Gene …-CTA-AAA-GAA-GGT-GTT-AGC-AAG-GTT-… Protein sequence …-L-K-E-G-V-S-K-D-… one amino acid Unfolded protein spontaneous self-organisation (~1 second) Native biologically relevant state not unique mobile inactive expanded irregular unique shape precisely ordered stable/functional globular/compact helices and sheets Structure-based informatics: De novo prediction of protein structure sample conformational space such that native-like conformations are found select hard to design functions that are not fooled by non-native conformations (“decoys”) astronomically large number of conformations 5 states/100 residues = 5100 = 1070 Semi-exhaustive segment-based folding EFDVILKAAGANKVAVIKAVRGATGLGLKEAKDLVESAPAALKEGVSKDDAEALKKALEEAGAEVEVK generate … Make random moves to optimise what is observed in known structures … Find the most protein-like structures minimise … … filter all-atom pairwise interactions, bad contacts compactness, secondary structure, consensus of generated conformations CASP prediction for T215 5.0 Å Cα RMSD for all 53 residues Ling-Hong Hung/Shing-Chung Ngan CASP prediction for T281 4.3 Å Cα RMSD for all 70 residues Ling-Hong Hung/Shing-Chung Ngan CASP prediction for T138 4.6 Å Cα RMSD for 84 residues CASP prediction for T146 5.6 Å Cα RMSD for 67 residues CASP prediction for T170 4.8 Å Cα RMSD for all 69 residues Structure-based informatics • Make predictions of peptides without the presence of substrates using de novo protocol • Make predictions of peptides in the presence of substrates using physics-based force-fields such as GROMACS • Analyse for similarity of structures (local and global) as well as common contact patterns between atoms in amino acids – the structural similarities and patterns give us the structural patterns responsible for folding and inorganic substrate binding • Perform higher-order simulations that involve many copies of a single or multiple peptides to generate sequences with specific stabilities and inorganic binding properties – larger assemblies for more controlled binding Computational design • Select the most promising candidate peptides generated from the sequence- and structure-based informatics for further simulation and design • Simulations can be performed to ensure that active sites and/or topologies found in nature are grafted onto these peptides • Experimental validation – synthesise peptides and check for binding activity • Main goal here is to help with rational design of inorganic binding peptides and focus experimental efforts in a more optimal manner • A good framework to obtain knowledge obtained experimentally with state of the protein structure prediction methodologies Grafting of biological active sites onto engineered peptides TIM barrel proteins 2246 with known structure hydrolase ligase lyase oxidoreductase transferase Acknowledgements Samudrala group: Aaron Chang Chuck Mader David Nickle Ekachai Jenwitheesuk Gong Cheng Jason McDermott Jeremy Horst Sarikaya group: Kai Wang Ling-Hong Hung Michal Guerquin Shing-Chung Ngan Stewart Moughon Tianyun Lu Zach Frazier Ersin Emre Oren National Institutes of Health National Science Foundation Searle Scholars Program (Kinship Foundation) Puget Sound Partners in Global Health UW Advanced Technology Initiative in Infectious Diseases http://bioverse.compbio.washington.edu http://protinfo.compbio.washington.edu