* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Extrachromosomal DNA wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
DNA barcoding wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic code wikipedia , lookup
Human genome wikipedia , lookup
Non-coding DNA wikipedia , lookup
Pathogenomics wikipedia , lookup
Microsatellite wikipedia , lookup
Genomic library wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome editing wikipedia , lookup
Point mutation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
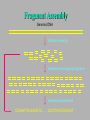
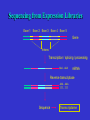
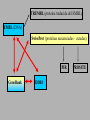
Bioinformática Bancos de datos biológicos Prof. Mirko Zimic [email protected] En 1944 IBM y la Universidad de Harvard estrenan Mark I, la primera computadora que responde a la moderna definición. Medía.15 metros de largo, 2.40 mts de alto y pesaba 10 toneladas. Utilizaba relays electromecánicos. Collecting Sequence Data • Genome (DNA-level): Genomic sequencing Complete picture of genome Generates physical map Includes regulatory and other silent regions • Transcriptome (RNA-level): Expression-library sequencing Expressed genes only Splicing / variant forms Can correlate with levels of expression • Proteome (protein-level): Protein sequencing Insight into biological function Gives information on protein-protein interactions Post-translational modifications detected DNA Sequencing DNA Sequencing (Cont’d) Fragment Assembly Genomic DNA Random shearing Sequence overlapping fragments Sequences assembled CCAGATTACGAAATCC . . . GGCTTATACCGGCAT Sequencing from Expression Libraries Exon 1 Exon 2 Exon 3 Exon 4 Exon 5 Gene Introns Transcription / splicing / processing AAA…AAA mRNA Reverse transcriptase AAA…AAA TTT…TTT Sequence Transcriptome Secuenciamiento de Proteínas Digital Storage of Sequence Data • Bit: A binary digit represented in a digital circuit; only two states recognized, 0 and 1 (usually 0 V and +5 V, respectively). • Byte: Grouping of 8 bits into a larger unit. Bits are usually numbered 0-7 (not 1-8!). • ASCII: Acronym for American Standard Code for Information Interchange. Representation of alphanumeric and some special characters as 1-byte (8 bit) unsigned integers {0 ... 255} (the set {20-1 ... 28-1}). The ASCII character set also includes nonprinting control characters such as carriage return (CR) or line feed (LF). Minimum storage requirement for human genome data represented as ASCII characters: 3109 bytes (3000 Mbytes) or about 5 CD-ROMs, exclusive of annotations or other data Number Systems Dec Bin Octal Hex Dec Bin Octal Hex 0 0 0 0 10 1010 12 A 1 1 1 1 11 1011 13 B 2 10 2 2 12 1100 14 C 3 11 3 3 13 1101 15 D 4 100 4 4 14 1110 16 E 5 101 5 5 15 1111 17 F 6 110 6 6 16 10000 20 10 7 111 7 7 17 10001 21 11 8 1000 10 8 18 10010 22 12 9 1001 11 9 19 10011 23 13 The ASCII Table Extended ASCII Characters Nucleic-acid Base Codes Symbol Meaning Symbol Meaning A A S G or C G G W A or T C C H A, C, or T (~G) T T B C, G, or T (~A) R A or G V A, C, or G (~T) Y C or T D A, G, or T (~C) M A or C N A, C, G, or T K G or T Adapted from Mount, Bioinformatics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (2001) Amino-acid Codes 1-letter Code 3-letter Code Amino Acid 1-letter Code 3-letter Code Amino Acid A Ala alanine N Asn asparagine C Cys cysteine P Pro proline D Asp aspartic acid Q Gln glutamine E Glu glutamic acid R Arg arginine F Phe phenylalanine S Ser serine G Gly glycine T Thr threonine H His histidine V Val valine I Ile isoleucine W Trp tryptophan K Lys lysine X Xxx undetermined L Leu leucine Y Tyr tyrosine M Met methionine Z Glx Glu or Gln Adapted from Mount, Bioinformatics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (2001) The exponential growth of molecular sequence databases & cpu power — Year BasePairs Sequences 1982 1983 1984 1985 1986 1987 1988 1989 1990 1991 1992 1993 1994 1995 1996 1997 1998 1999 2000 2001 680338 606 2274029 2427 3368765 4175 5204420 5700 9615371 9978 15514776 14584 23800000 20579 34762585 28791 49179285 39533 71947426 55627 101008486 78608 157152442 143492 217102462 215273 384939485 555694 651972984 1021211 1160300687 1765847 2008761784 2837897 3841163011 4864570 11101066288 10106023 14396883064 13602262 doubling time ~ one year What are sequence databases? These databases are an organized way to store the tremendous amount of sequence information accumulating worldwide. Most have their own specific format. North America: the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM), at the National Institute of Health (NIH), has GenBank & GenPept. Europe: the European Molecular Biology Laboratory (EMBL), the European Bioinformatics Institute (EBI), and the Swiss Institute of Bioinformatics’ (SIB) Expert Protein Analysis System (ExPasy), all help maintain the EMBL Nucleotide Sequence Database, and the SWISSPROT & TrEMBL amino acid sequence databases. Asia: The National Institute of Genetics (NIG) supports the Center for Information Biology’s (CIG) DNA Data Bank of Japan (DDBJ). More organization stuff — Nucleic acid sequence databases (and TrEMBL) are split into subdivisions based on taxonomy (historical rankings — the Fungi warning!). PIR is split into subdivisions based on level of annotation. TrEMBL sequences are merged into SWISS-PROT as they receive increased levels of annotation. • Nucleic Acid DB’s – GenBank/EMBL/DDBJ • all Taxonomic categories • “Tags” – EST’s – GSS’s • Amino Acid DB’s – SWISS-PROT • TrEMBL – PIR • PIR1 • PIR2 • PIR3 • PIR4 • NRL_3D – Genpept • TrEMBL contains the translations of all coding sequences (CDS) present in the EMBL Nucleotide Sequence Database, which are not yet integrated into SwissProt. • PIR (Protein Information Resource) produces the Protein Sequence Database (PSD) of functionally annotated protein sequences, which grew out of the Atlas of Protein Sequence and Structure (1965-1978) edited by Margaret Dayhoff TREMBL (proteina traducida del EMBL) EMBL (DNA) SwissProt (proteínas secuenciadas – curadas) PIR GeneBank DDBJ PROSITE What about other types of biological databases? • Three dimensional structure databases: • the Protein Data Bank and Rutgers Nucleic Acid Database. • These databases contain all of the 3D atomic coordinate data necessary to define the tertiary shape of a particular biological molecule. The data is usually experimentally derived, either by X-ray crystallography or with NMR, but sometimes it is a hypothetical model. In all cases the source of the structure and its resolution is clearly indicated. • Secondary structure boundaries, sequence data, and reference information are often associated with the coordinate data, but it is the 3D data that really matters, not the annotation. Other types of Biological DB’s — • Still more; these can be considered ‘non-molecular’: • Genomic linkage mapping databases for most large genome projects (w/ pointers to sequences) — H. sapiens, Mus, Drosophila, C. elegans, Saccharomyces, Arabidopsis, E. coli, . . . . • Reference Databases (also w/ pointers to sequences): e.g. • OMIM — Online Mendelian Inheritance in Man • PubMed/MedLine — over 11 million citations from more than 4 thousand bio/medical scientific journals. • • Phylogenetic Tree Databases: e.g. the Tree of Life. • Metabolic Pathway Databases: e.g. WIT (What Is There) and Japan’s GenomeNet KEGG (the Kyoto Encyclopedia of Genes and Genomes). • Population studies data — which strains, where, etc. And then databases that many biocomputing people don’t even usually consider: • e.g. GIS/GPS/remote sensing data, medical records, census counts, mortality and birth rates . . . . Large Databases • Once upon a time, GenBank sent out sequence updates on CD-ROM disks a few times per year. • Now GenBank is over 40 Gigabytes (11 billion bases) • Most biocomputing sites update their copy of GenBank every day over the internet. • Scientists access GenBank directly over the Web Finding Genes in GenBank •These billions of G, A, T, and C letters would be almost useless without descriptions of what genes they contain, the organisms they come from, etc. •All of this information is contained in the "annotation" part of each sequence record. Entrez is a Tool for Finding Sequences • GenBank is managed by the NCBI (National Center for Biotechnology Information) which is a part of the US National Library of Medicine. • NCBI has created a Web-based tool called Entrez for finding sequences in GenBank. http://www.ncbi.nlm.nih.gov • Each sequence in GenBank has a unique “accession number”. • Entrez can also search for keywords such as gene names, protein names, and the names of orgainisms or biological functions Entrez is Internally Cross-linked • DNA and protein sequences are linked to other similar sequences • Medline citations are linked to other citations that contain similar keywords • 3-D structures are linked to similar structures Databases contain more than just DNA & protein sequences Proyecto Genoma Humano La secuencia del genoma está casi completa! – aproximadamente 3.5 billones de pares de bases. Raw Genome Data Gene finding Data Quality Issues Bioinformatics Databases • • • • Usually organised in flat files Huge collection of Data Include alpha-numeric and pictorial data Latest databases have gene/protein expression data (images) Demand • High quality curated data • Interconnectivity between data sets • Fast and accurate data retrieval tools – queries using fussy logic • Excellent Data mining tools – For sequence and structural patters Errors in DNA sequence and Data Annotation • Current technology should reduce error rates to as low as 1 base in 10000 as every base is sequenced between 6-10 times and at least one reading per strand. • Therefore, in a procaryote, error of 1 isolated wrong base would result to one amino acid error in ~10-15 proteins. • In human genome gene-dense regions contain about 1 gene per 10000 bases, with average estimated at 1 gene per 30000bases. • Therefore, corresponding error rate would be roughly one amino acid substitution in 100 proteins. • But large scale error in sequence assembly can also occur. Missing a nucleotide can cause a frameshift error. DNA data … • The DNA databases (EMBL/ GenBank/ DDBJ) carry out quality checks on every sequence submitted. • No general quality control algorithm is yet in widespread use. • Some annotations are hypothetical because they are inferences derived from the sequences. – Ex. Identification of coding regions. These inferences have error rates of their own. DNA Sequencing (Cont’d)