* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Translation
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Catalytic triad wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Butyric acid wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Gene expression wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Proteolysis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Messenger RNA wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Epitranscriptome wikipedia , lookup
Genetic code wikipedia , lookup
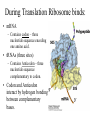
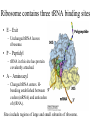
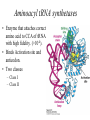
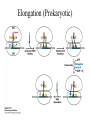
Biochemistry I (CHE 418 / 5418) Reading Assignment Berg et. al (2007) Chapter 30 Central Dogma of Molecular Biology Replication DNA Transcription RNA Translation Protein mRNA. tRNA, rRNA • Replication - DNA directed DNA synthesis • Transcription - DNA directed RNA synthesis • Translation - RNA directed Protein synthesis Translation RNA directed protein synthesis – Four letters of nucleic acid language translated into 20 amino acids of protein language. • mRNA, tRNA, ribosome (rRNA) • Occurs on ribosome. • Protein synthesized in N (amino) to C (carboxyl) direction • mRNA read from 5’ to 3’ N 5’ N N N 3’ Translation Divided into 3 Stages Initiation Elongation Ribosome binds mRNA identifies start codon (AUG). Termination Protein synthesized N to C Polypeptide released upon encountering stop codons (UAA, UAG, UGA) * Met in Eukaryotes * fMet N-formylmethionine in prokaryotes During Translation Ribosome binds: • mRNA – Contains codon – three nucleotide sequence encoding one amino acid. • tRNAs (three sites) – Contains Anticodon – three nucleotide sequence complementary to codon. • Codon and Anticodon interact by hydrogen bonding between complementary bases. Ribosome contains three tRNA binding sites • E – Exit – Uncharged tRNA leaves ribosome. • P – Peptidyl – tRNA in this site has protein covalently attached. • A – Aminoacyl – Charged tRNA enters. Hbonding established between codon (mRNA) and anticodon of (tRNA). Sites include regions of large and small subunits of ribosome. Reaction to Add Amino Acids • Amino acids are added to the C terminus by transferring existing chain to next amino acid. Polysome (Polyribosome) • Polysome -multiple ribosomes bond to the same mRNA. – Each ribosome in a polysome is synthesizing the same protein. • In Prokayotes, transcription and translation may be coupled, since both are occurring in the same compartment at the same time. tRNA carry amino acids • Amino acid (C terminus) attached via an ester to 2’or 3’ hydroxyl of A of CCA on tRNA. • “Charged tRNA” – tRNA with correct amino acid attached. • Aminoacyl tRNA synthetases – class of enzymes that attach amino acids to tRNA. Aminoacyl tRNA synthetases • Enzyme that attaches correct amino acid to CCA of tRNA with high fidelity. (>10-4). • Binds Activation site and anticodon. • Two classes – Class I – Class II Two Classes of Aminoacyl tRNA synthetases • Two classes differ in: – Sequence alignment / comparison. • 10 tRNA per class. – Binding to tRNA on different “face” or side – Attaching amino acid to different hydroxyl • Class 1 to 2’ OH • Class 2 to 3 ‘ OH – Binding ATP in different comformation – Subunit comformation • Class 1 are monomers • Class II are dimers Aminoacyl tRNA synthetases reaction • Two step reaction – 1. AA + ATP ↔ Aminoacyl-AMP + PPi • Amino acid attached to AMP via mixed anhydride which conserves the energy of phosphodieaster bond in ATP. • Acyl adenylate – 2. Amino acid transferred to tRNA. • Ester linkage to 2’ or 3’ hydroxyl of A of CCA on 3’ end of tRNA. Threonyl-tRNA synthetase An example of Fidelity • How does Threonyl-tRNA synthetase select between similarly shaped amino acids? • Thr has almost the same shape as Ser and Val. – 1. Active site selection – 2. Editing site Threonyl-tRNA synthetase An example of Fidelity (Cont’) • 1. Active site selection by selective binding of substrate. – Zn 2+ • Coordinated to enzyme by 2 his and 1 cys residues • Binds Thr by amino group and OH of side chain – Asp of enzyme • Binds OH of thr substrate. • This interaction prohibits attachment of Val. Threonyl-tRNA synthetase An example of Fidelity (Cont’) • 2. Editing site – Removes Mischarged amino acids (Ser). • CCA with amino acid attached swings into editing site. Product does not dissociate before editing. • Increasing fidelity to > 10-4 NOT ALL Aminoacyl tRNA synthetases require editing. Codon hydrogen bonds to Anticodon • Codon of mRNA and Anticodon of tRNA are antiparellel. Wobble • Third base of anticodon (5’ end) allows binding with multiple bases. Codon Usage Table • mRNA are “read” three nucleotides (codon) at a time starting from a fixed point. The Genetic code is unambiguous, degenerate, non-overlapping and universal • Unambiguous – A given codon either designates a single amino acid or is a stop codon. • Degenerate – more than one codon can specify the same amino acid, so the genetic code is said to be degenerate. • Non-overlapping – the code is read sequentially, one codon after another without spacer bases, from a fixed starting point. • Universal Initiation (Prokaryotic) • Prokaryotic start codon = AUG. – Others exist (rare) • GUG = Val • UUG = Leu • AUG encodes fMET • Two important interactions – 1. Binding of mRNA to 3’ end of 16S rRNA. – 2. H bonding of initiator codon with anticodon. • tRNAf binds to P site of ribosome. Elongation (Prokaryotic) • P site is occupied by tRNA holding fMet, or later the growing polypeptide • Elongation factors bring the correct charged tRNA to the A site. – EF-Tu – brings charged tRNA to A site of ribosome. Requires bond GTP (hydrolyzed to GDP during reaction). GTP binds to p-loop. – EF-Ts – removes GDP from EF-Tu and replaces with GTP • The 23 S rRNA of the ribosome catalyzes the attachment of amino acids. • Translocation is accelerated by EF-G. Elongation (Prokaryotic) Termination (Prokaryotic) • Termination aided by RF = release factors. Castor Oil • What is Castor Oil? – Thick, yellowish or almost colorless oil extracted from castor bean (seed). • Uses of Castor Oil – – – – – – – Laxative Lubricating oil Quick-drying oil (when dehydrated) used in paints and varnishes. Competes with linseed and tung oil Coating fabrics /protective coverings Sebacic acid - basic ingredient in the production of nylon 6, 10 and other synthetic resins / fibers Castor Bean • Ricinus communis – (family Euphorbiaceae) • Source of Castor Oil http://www.ansci.cornell.edu/plants/alphalist.html http://waynesword.palomar.edu/plmar99.htm#flow Synthesis of Nylon 6, 10 – Condensation Polymerization – Sebacic acid + 1, 6-hexanediamine – Sebacic acid – 10-carbon dicarboxylic acid – carboxylic group (COOH) at each end of the molecule – 1,6-hexanediamine – 6-carbon molecule with an amino group (CNH2) at each end. – Free carboxylic acid of Sebacic Acid reacts with amino group of 1, 6hexamediamine – water molecule is produced at each link. What is Ricin? • Protein (64 kda) found in pulp from production of castor oil. – Heterodimer • A chain – 267 amino acid residues. – Inhibits protein synthesis. • B-chain – 262 amino acid residues. – facilitates transport into cell. • Connected by disulfide bridge. Mode of Action of Ricin • A chain – N-glycosidase which removes specific adenine (depurinates) 28 S ribosomal RNA • A-4323 in rat liver – prevents the binding of an elongation factor, leaving the ribosome incapable of protein synthesis. – Extremely toxic • 70 micrograms (equivalent to weight of grain of salt) will kill a 160 pound man. – Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals (1997) Many Antibiotics and Toxins Inhibit Translation