* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PowerPoint - Biological Sciences
Silencer (genetics) wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Paracrine signalling wikipedia , lookup
Peptide synthesis wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Point mutation wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene expression wikipedia , lookup
Epitranscriptome wikipedia , lookup
Expression vector wikipedia , lookup
Signal transduction wikipedia , lookup
Structural alignment wikipedia , lookup
Interactome wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Metalloprotein wikipedia , lookup
Homology modeling wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Protein Structure 2
Protein Modification
BL4010 10.03.06
Nomenclature
• Domain - a particular region within a
polypeptide chain (ATP binding domain)
• Motif - a protein structural element (may
appear more than once in a single protein or
in many different types of proteins (e.g. greek
key)
• Subunit - a single polypeptide unit within a
larger multipeptide protein
• Complex - group of proteins with long-term or
transient physical association
Multidomain proteins
Multisubunit proteins
Quaternary (4°) structure
What are the forces driving quaternary association?
• Typical Kd for two subunits: 10-8 to 10-16M!
• These values correspond to energies of 50-100
kJ/mol at 37 C
• Entropy loss due to association - unfavorable
• Entropy gain due to burying of hydrophobic
groups - very favorable!
Alcohol dehydrogenase dimer
Prealbumin dimer
Tyrosine kinases
Packing symmetry
Multiple subunits vs. multiple domains
• Structural factors
–
–
–
–
Stability: reduction of surface to volume ratio
Bringing catalytic sites together (efficiency)
Flexibility in shared binding sites
Increased complexity
• Genetic factors
– Gene duplication = genetic freedom/economy
– Multiple interactions = increased impact of
mutation
• Regulation
– Cooperativity
– Must be co-expressed
Tubulin as an
example
• Essential component of
the cytoskeleton
• Dimers polymerize
• Multiple sites of
interaction
• Dynamic
• Mutations have severe
effects
Protein Modification
Post-translational Modifications
•
•
•
•
•
•
•
•
•
•
Cleavage of signal peptides
Phosphorylation
Amidation
Glycosylation
Hydroxylation
Ubiquitination
Addition of prosthetic groups
Iodination
Adenylation
Sulfonation
• Prenylation
• Myristoylation
• Acylation
– Acetylation
– Methylation
• Oxidative crosslinking
• N-Glutamyl cyclization
• Carboxylation
Table 4.1
Are disulfide bonds posttranslational modification?
N-terminal modifications
• N-formyl Met (by product of translation
process and incomplete cleavage by
deformylase)
• Aminopeptidase cleavage of Met
• N-terminal acetylation
• N-terminal myristoylation
• N-terminal glutamyl cyclization
C-terminal modification
• C-terminal prenylation
– farnesylation
– geranylation/geranyl-geranylation
• C-terminal amidation
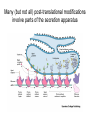
Many (but not all) post-translational modifications
involve parts of the secretion apparatus
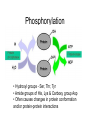
Phosphorylation
• Hydroxyl groups - Ser, Thr, Tyr
• Amide groups of His, Lys & Carboxy group Asp
• Often causes changes in protein conformation
and/or protein-protein interactions
C-terminal Amidation
• Neuropeptides and
hormones
•
•
C-terminal glycine is hydroxylated
alpha-hydroxy-glycine.
Glyoxylate. C-terminal amidation is
essential to the biological activity of
many neuropeptides and hormones.
Hydroxylation and
Carboxylation
• Hydroxylation (previous lecture)
– Lys and Pro (e.g. collagen)
– Requires vitamin C
• Carboxylation
– Requires viatamin K
Iodination
• Special case: Tyr thyroxin
Protein Acylation
• Methylation and acetylation
– acetylation of histone Lys
• S-acylation (Cys)
• Prenylation (C-terminus)
• Myristoylation (N-terminus)
Acetylation
N-terminal acetylation
• No simple sequence consensus
• Usually accompanied by cleavage of
the N-terminal Met
Mass spectral analysis
Methylation
Can occur at carboxyls, amides, sulfhydryl, etc.
SAM is the activated carbon source
Multiple methylation
Histones
• DNA binding
proteins
• Chromatin
structure
• Affect gene
expression
Sulfonation
• O-sulfonation of Ser/Thr discovered in
2004 by MS analysis (Mol Cell
Proteomics. 3(5):429-40)
N-terminal myristoylation
• Consensus Gly-X-X-X-Ser/Thr
myristoyl Coenzyme A
methylated c-terminus
C-terminal
Prenylation
Prenylation
CAAX motif
AAX cleaved and Cys methylated then prenyl
group added
n-terminus
by thioether linkage (C-S-C).
S-acylation
Adenylation
• Toxins frequently adenylate host target
proteins to inactivate them
• Glutamine synthetase is inactivated by
adenylylation and activated by
deadenylylation
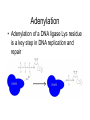
Adenylation
• Adenylation of a DNA ligase Lys residue
is a key step in DNA replication and
repair
Proteolytic cleavage
All non-cytoplasmic proteins must be translocated
• The leader peptide retards the folding of the
protein so that molecular chaperone proteins
can interact with it and direct its folding
• The leader peptide also provides recognition
signals for the translocation machinery
• A leader peptidase removes the leader
sequence when folding and targeting are
assured
Proteolytic processing
• Proteolytic cleavage of the hydrophobic Nterminal signal peptide sequence
• Proteolytic cleavage at a site defined by pairs
of basic amino acid residues
• Proteolytic cleavage at sites designated by
single Arg residues
• Signal prediction algorithms (e.g. SignalP)
caution: may predict TMDs as signalPs
Hormones
• All secreted polypeptide hormones are
synthesized with a signal sequence (which
directs them to secretory granules, then out)
• Usually synthesized as inactive
preprohormones ("pre-pro" implies at least
two processing steps)
• Proteolytic processing produces the
prohormone and the hormone
GASTRIN
• Product of preprogastrin
– 101-104 residues
• Signal peptide cleavage
– prepropeptide propeptide 80-83 residues
• Cleavage at Lys and Arg residues and C-terminal
amidation leaves gastrin
– propeptide peptide
• N-terminal residue of gastrin is pyroglutamate
• C-terminal amidation involves destruction of Gly
GASTRIN
GASTRIN
Heptadecapeptide secreted by the antral mucosa of stomach
stimulates acid secretion in stomach
Glycosylation: 2 flavors
N-linked GlcNAc
(Asn)
O-linked GalNac
(Ser, Thr, Hyp)
Glycosylation
Protein glycosylation
• Usually extracellular or at cell surface
• High structural information content
– molecular recognition
• Occurs along the secretory pathway
• Often stabilizes structure
• Difficult to get crystal structure for more
than one or two carbohydrate residues
Protein glycosylation
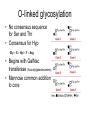
O-linked glycosylation
• No consensus sequence
for Ser and Thr
• Consensus for Hyp
Gly – X – Hyl – Y – Arg
• Begins with GalNac
transferase (N-acetylgalactosamine)
• Mannose common addition
to core
The ABO(H) blood group determinant is
an O-linked polysaccharide
•In 1901, Karl Landsteiner
discovered blood group
antigens.
•Since that time nearly
200 antigens have been
identified.
•Carbohydrate-dependant
blood group antigens are
carried by both
glycoproteins and
glycolipids
N-linked glycosylation
• Most proteins contain several potential
sites but usually only a few (in any) are
actually glycosylated
• Many different patterns
• Consensus (-N-X-T/S-)
Copyright 2000-2002
IonSource.Com
N-linked glycosylation begins
in the cytoplasm
glycosyl-dolichol-phosphate
(lipid) core is transferred to protein
Further processing and elaboration
occurs in the ER and Golgi
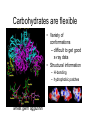
Carbohydrates are flexible
• Variety of
conformations
– difficult to get good
x-ray data
• Structural information
– H-bonding
– hydrophobic patches
wheat germ agglutinin
GPI anchors
(glycosylphosphatidyl inositol)
Carbohydrates as cross-linkers
Galectin-1 dimer
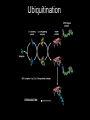
Ubiquitination
DEGRADATION
Ubiquitination
• Ubiquitinating enzymes E1,E2, E3 thiol ester bond
• Final target - isopeptide bond between
a lysine residue of the substrate (or the
N terminus of the substrate) and
ubiquitin
• Ubiquitin first activated by adenylation
• Science 2005 309:127-130 target
protein ubiquitination on Cys!
More about proteoglycans and
ubiquitination later