* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download GCE Biology BY5 1075-01
Genome (book) wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
DNA barcoding wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Genome evolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genetically modified food wikipedia , lookup
Gene expression programming wikipedia , lookup
Population genetics wikipedia , lookup
Genetic drift wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Genetically modified crops wikipedia , lookup
Koinophilia wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Genetic engineering wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Designer baby wikipedia , lookup
Helitron (biology) wikipedia , lookup
History of genetic engineering wikipedia , lookup
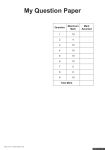
WJEC 2014 Online Exam Review GCE Biology BY5 1075-01 All Candidates' performance across questions Question Title 1 2 3 4 5 6 7 8 N 5207 5207 5211 5210 5192 5207 5202 5194 Mean 4.4 2.9 5.2 6.8 6.2 6 6.6 4.5 SD 1.9 1.5 1.6 2.5 3.1 2 2.8 2.1 Max Mark 8 6 8 11 12 12 13 10 FF 55.5 47.8 65 61.7 51.7 50.1 51 44.8 Attempt % 99.9 99.9 100 99.9 99.6 99.9 99.8 99.6 GCE Biology BY5 1075-01 8 44.8 7 51 Question 6 50.1 5 51.7 4 61.7 3 65 2 47.8 1 55.5 0 10 20 30 40 50 60 Facility Factor % 70 80 90 100 5 6 7 14 5. The techniques of recombinant DNA technology and micro-propagation are used to produce Genetically Modified Crops. The following summary is adapted from an account given on the Food Standards Agency’s web site [www.food.gov.uk] 1. A plant with the desired characteristic is identified – e.g. resistance to the herbicide ‘Roundup’. 2. The specific gene that produces this characteristic is found in the plant’s DNA and cut out. 3. To get the gene into the cells of the plant being modified, the gene needs to be attached to a carrier. A piece of bacterial DNA called a plasmid is joined to the gene to act as the carrier. 4. Once the gene is attached to the plasmid, a marker gene is also added to identify which plant cells take up the new gene. 5. 6. A copy of the ‘gene package’ is dried onto a gold or tungsten particle – and fired into a piece of tissue from the plant being modified. The particle carries the ‘gene package’ into the plant’s cells. 7. The plant tissue is put into a selective growth medium so that only modified tissue develops into plants. (a) Explain how different types of enzymes are used in stages 2 and 3 to produce the ‘gene package’.[4] The ‘gene package’ is put in a bacterium, which multiplies, to create many copies of the ‘gene package’. © WJEC CBAC Ltd. (1075-01) Examiner only 15 (b) Describe the steps involved in the culture of a large number of genetically identical plants from the plant tissue produced in stage 7. [3] Examiner only (i) Explain the advantage to farmers of having crops resistant to ‘Roundup’. (c) [3] (ii) Explain why environmentalists might have legitimate objections to using GM crops resistant to ‘Roundup’. [2] 12 © WJEC CBAC Ltd. (1075-01) Turn over. 17 6. A species of mouse Peromyscus polionotus found in Florida, USA, has a number of different coat colours. Coat colour in mice is controlled by several genes. Dark fur is produced when the hair producing cells secrete a pigment called eumelanin. A high level of eumelanin is produced when a transmembrane protein called MC1R is stimulated by a hormone. (a) The diagram below shows part of the amino acid sequence of MC1R, part of the sequence of nucleotides in the gene for MC1R and how it might change to produce light fur: Original Amino acid sequence Nucleotide sequence (allele R) lle Thr Lys Asn Arg Asn Leu His Ser ATCACCAAAAACCGCAACCTGCACTCG Changed to produce light fur Amino acid sequence Nucleotide sequence (allele C) lle Thr Lys Asn Cys Asn Leu His Ser ATCACCAAAAACTGCAACCTGCACTCG (i) Describe the change in the gene and the subsequent change in the MC1R molecule.[2] (ii) Using the information provided, explain how this change results in mice with light fur.[2] © WJEC CBAC Ltd. (1075-01) Turn over. Examiner only 18 (b) This change in the MC1R gene means that there are two alleles, R and C. The map below shows the distribution of the different coloured mice and the relative frequencies of the alleles R and C in each population. Northern Florida sandy beaches forest/scrub Allele frequency 5% R 95% C Population 3 Allele frequency 30% R 70% C Population 2 Allele frequency 100% R 0% C Population 1 (i) Use the diagram to suggest how fur colour is related to environmental conditions. [2] (ii) Under what circumstance could the difference between the allele frequencies in populations 2 and 3 be explained by genetic drift, despite both living on beaches?[1] © WJEC CBAC Ltd. (1075-01) Examiner only 19 (iii) Explain how Natural Selection could have caused the relative allele frequency shown in population 3.[4] Examiner only (iv) Under what circumstances would the mouse population become a separate species?[1] 12 © WJEC CBAC Ltd. (1075-01) Turn over. 19 Examiner only 7. The following is a quotation from an ecological investigation. “Lowland heaths are high-profile ecosystems for conservation action in England, but they are under threat from invasion by Betula spp., Pinus sylvestris, and Ulex europaeus.” [R.J. Mitchel et al. Journal of Applied Ecology, 1997, 37, 1426-1444] (a) Distinguish between primary succession and secondary succession. © WJEC CBAC Ltd. (1075-01) [2] Turn over. 20 The authors studied a number of heathland sites in Dorset including Arne, Blackhill, and Higher Hyde, where succession to one or another of the three species had taken place. The data below are based on the paper but have been simplified and modified for illustrative purposes. The successional stages in the study were named according to the dominant invasive species; plus B, where Betula spp, was the invader, plus PS, where Pinus sylvestris was the invader and plus U, where Ulex europaeus, was the invader. (b) The group examined changes in soil chemistry from the original heath stage. Some of their results are summarised in the table below: soil chemical property value by succession stage original heath plus B plus PS plus U Arne 3.63 4.01 3.60 3.63 Blackhill 3.52 3.66 3.48 3.54 Higher Hyde 3.53 5.06 3.51 3.47 3.56 4.24 3.53 3.55 Arne 2.41 3.85 2.69 3.16 Blackhill 4.15 4.91 3.79 4.55 Higher Hyde 5.08 5.35 3.55 4.76 3.88 4.70 3.34 4.16 Arne 0.51 0.65 0.59 1.16 Blackhill 0.84 0.88 0.97 2.31 Higher Hyde 0.69 0.98 1.17 3.64 0.68 0.84 0.91 2.37 pH mean phosphorus μgPg –1 mean nitrate/nitrite μgNg –1 mean (i) What do the pH values tell us about the soil in all stages in all sites? [1] .................................................................................................. (ii)Use mean values from the table above to compare three changes to soil chemistry following invasion by Betula spp. with the changes following invasion by Ulex europaeus.[3] pH . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . phosphorus nitrate/nitrite ................................................................................................................................................................ .............................................................................................................................................................. © WJEC CBAC Ltd. (1075-01) Examiner only 21 Examiner only (c) The table below shows changes to the vegetation in the successional stages: Species (by successional stage) % cover of species (by site) Arne Blackhill Higher Hyde original heath Calluna vulgaris Erica cinerea Erica tetralix Cladonia portentosa 62.0 22.4 9.9 8.5 66.1 25.7 2.6 0 88.2 2.6 9.9 0.5 plus B Betula spp. Agrostis curtisii Pteridium aquilinum Calluna vulgaris 18.9 0.0 25.2 0.0 11.7 53.6 7.5 0.0 16.5 0.0 1.6 0.4 plus PS Pinus sylvestris Pteridium aquilinum Erica cinerea Calluna vulgaris 36.2 0.3 0.0 0.0 38.2 24.7 0.0 0.0 plus U Ulex europaeus Calluna vulgaris Erica cinerea Erica tetralix 87.0 14.7 1.5 0.1 75.3 5.8 11.3 0.3 79.0 7.2 4.3 0.3 (i) Which invading species has least impact on the vegetation on the original heathland?[1] (ii) With reference to the data for plus B in both tables suggest a mechanism by which changes to vegetation occur during succession. [2] © WJEC CBAC Ltd. (1075-01) Turn over. 22 (d) Sixteen years later some of these successions have reached their natural conclusions. (i) Examiner only What name is given to the group of organisms that inhabit the ecosystem at the end [1] of successional change? (ii) What usually happens to species diversity as succession proceeds? [1] (iii)Using named species from the table in part (c) explain why conservationists in Dorset are taking steps to prevent plus B and plus PS succession in heathland, but are less worried about type plus U succession. [2] 13 © WJEC CBAC Ltd. (1075-01)