* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 6-Catabolism of Pyrimidine Nucleotides
Genetic code wikipedia , lookup
Lipid signaling wikipedia , lookup
Epitranscriptome wikipedia , lookup
Proteolysis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Polyadenylation wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Biochemistry wikipedia , lookup
Peptide synthesis wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
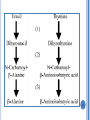
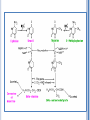
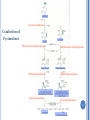
CATABOLISM OF PYRIMIDINE NUCLEOTIDES 1 Dr. Shumaila Asim Lecture # 6 PYRIMIDINE CATABOLISM Pyrimidines are catabolized to β-alanine and βaminoisobutyrate then secreted in urine. 2 Degradation of pyrimidine nucleotide The pyrimidine ring can be completely degraded in humans. The products include: NH3, CO2, b-alanine, and baminoisobutyrate. Both b-alanine, and b-aminoisobutyrate can be further converted into acetyl-CoA and succinyl-CoA, respectively, or are excreted in the urine. 4 5 Cytosine deaminase Catabolism of Pyrimidines dihydrouracil dehydrogenase dihydrouracil dehydrogenase dihydropyrimidinase dihydropyrimidinase -ureidopropionase -ureidopropionase 6 7 PYRIMIDINE RELATED DISORDERS Orotic acidurias Deficiency of enzymes of urea cycle results in excretion of pyrimidine precursors 8 PYRIMIDINE RELATED DISORDERS Orotic acidurias Type – I Deficiency of both orotate phospho-ribosyl transferase and orotidylate decarboxylase Orotate & OMP cannot be converted to UMP ,CMP & TMP Orotate & OMP accumulate inhibition of DNA & RNA synthesis Type – II Deficiency of only orotidylate decarboxylase OMP cannot be converted into UMP inhibition of DNA / RNA synthesis 9 Pyrimidine Related Disorders Deficiency of Urea Cycle enzymes e.g. Deficiency of Ornithine trans-carbamoylase Carbamoyl-phosphate cannot enter urea cycle Exits into cytosol & stimulates pyrimidine synthesis Leads to increased excretion of precursors of pyrimidines i.e. Orotic acid, uracil and orotidine 10 POINTS Synthesis of Purine Nucleotides De novo synthesis: Site, Characteristics, Element sources of purine bases Salvage pathway: definition, significance, enzyme, Lesch-Nyhan syndrome Formation of deoxyribonucleotide: NDP level Antimetabolites of purine nucleotides: Degradation of Purine Nucleotides Uric acid, gout Synthesis of Pyrimidine Nucleotides De novo synthesis: Characteristics, Element sources of pyrimidine bases Salvage pathway Antimetabolites of pyrimidine nucleotides Purine, Amino acid, and Folic acid analogs Catabolism of Pyrimidine Nucleotides 11 1.The key substance in the synthesis of purine, phosphoribosyl pyrophosphate is formed by (A) ribose 5-phosphate (B) 5-phospho -ribosylamine (C) D-ribose (D) Deoxyribose 2. In purine biosynthesis ring closure in the molecule formyl glycinamide ribosyl-5-phosphate requires the cofactors: (A) ADP (B) NAD (C) FAD (D) ATP and Mg++ 3. Ring closure of formimidoimidazole carboxamide ribosyl-5-phosphate yields the first purine nucleotide: (A) AMP (B) IMP 12 (C) XMP (D) GMP 4. The cofactors required for synthesis of adenylosuccinate are (A)ATP, Mg++ (B) ADP (C) GTP, Mg++ (D) GDP 5. Conversion of inosine monophosphate to xanthine monophosphate is catalysed by (A) IMP dehydrogenase (B) Formyl transferase (C) Xanthine-guanine phosphoribosyl transferase (D) Adenine phosphoribosyl transferase 6. Phosphorylation of adenosine to AMP is catalysed by (A) Adenosine kinase (B) Deoxycytidine kinase (C) Adenylosuccinase (D) Adenylosuccinate synthetase 13 7. Pyrimidine and purine nucleoside biosynthesis share a common precursor: (A) PRPP (B) Glycine (C) Fumarate (D) Alanine 8.The first true pyrimidine ribonucleotide synthesized is (A) UMP (B) UDP (C) TMP (D) CTP 9. Methotrexate blocks the synthesis of thymidine monophosphate by inhibiting the activity of the enzyme: (A) Dihydrofolate reductase (B) Orotate phosphoribosyl transferase (C) Ribonucleotide reductase (D) Dihydroorotase 14