* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 20.Human.Neanderthal.Selection
Genomic imprinting wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetics and archaeogenetics of South Asia wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Population genetics wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Genetic drift wikipedia , lookup
Microevolution wikipedia , lookup
Human genetic variation wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Recent African origin of modern humans wikipedia , lookup
SNP genotyping wikipedia , lookup
Dominance (genetics) wikipedia , lookup
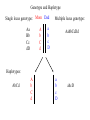
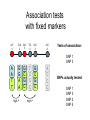
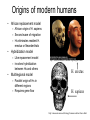
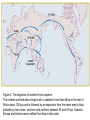
Rates of recombination around genes Most traits in organisms Show continuous variation How do we find the genes That affect these “quantitative” traits Scan the genome for Nucleotide sites that Co-vary with the phenotype Marker association studies Mutation “causing” variation in height Tall Tall Tall Tall Short Short Short Short A A A A G G G G Adjacent SNPs are linked Distant sites show no genotype-phenotype association Problem: how do we find the causal SNPs? Needle in a haystack Genotype and Haplotype Single locus genotype: Mom Dad Aa Bb Cc dD A b C d Multiple locus genotype: a b c D AabbCcDd Haplotypes: AbCd A b C d a b c D abcD Association tests with fixed markers A/T 1 G/A 2 G/C 3 T/C 4 G/C 5 A/C 6 Tests of association: SNP 1 SNP 3 A A T T G G A A high r2 G C G C T C C C high r2 G C G C A C C C SNPs actually tested: SNP 1 SNP 3 SNP 2 SNP 5 Origins of modern humans • African replacement model – African origin of H. sapiens – Second wave of migration – Hs eliminates resident H. erectus or Neanderthals • Hybridization model – Like repacement model – involves hybridization between Hs and others • Multiregional model – Parallel origin of Hs in different regions – Requires gene flow http://www.linfield.edu/~mrobert/originsfigure5.html H. erectus H. sapiens http://emuseum.mnsu.edu/biology/humanevolution/fosrec.html Figure 3. The migration of modern Homo sapiens. The scheme outlined above begins with a radiation from East Africa to the rest of Africa about 100 kya and is followed by an expansion from the same area to Asia, probably by two routes, southern and northern between 60 and 40 kya. Oceania, Europe and America were settled from Asia in that order. The common variant G6PD-202A confers partial protection against malaria, with a case-control study estimating a reduction in disease risk of about 50% The CD40 ligand gene (TNFSF5) encodes a proteinwith a critical role in immune response to infectious agents. TNFSF5-726C, is associated with a similar degree of protection against malaria. Extended haplotype homozygosity Sabeti et al. 2002: We measured LD at a distance x from the core region by calculating the extended haplotype homozygosity (EHH).EHHis defined as the probability that two randomly chosen chromosomes carrying the core haplotype of interest are identical by descent (as assayed by homozygosity at all SNPs8) for the entire interval from the core region to the point x. EHH thus detects the transmission of an extended haplotype without recombination. Our test for positive selection involves finding a core haplotype with a combination of high frequency and high EHH Core haplotypes and EHH Data: 800,000 polymorphic SNPs 309 unrelated individuals: • 89 Japanese and Han Chinese from Tokyo and Beijing denoted as East Asian (ASN) • 60 northern and western Europeans (CEU) • 60 Yoruba (YRI) from Ibadan, Nigeria. In plots of EHH versus distance, the area under the EHH curve will usually be much greater for a selected allele than for a neutral allele. We compute the integral of the observed decay of EHH away from a specified core allele until EHH reaches 0.05. This integrated EHH (iHH) denoted iHHA or iHHD, depending on whether it is computed with respect to the ancestral or derived core allele. In neutral models, low frequency alleles are younger and are associated with longer haplotypes than higher frequency alleles We adjust the unstandardized iHS to obtain our final statistic which has mean 0 and variance 1 regardless of allele frequency at the core SNP: Discovering the patterns of human migration from haplotypes Hominid fossil ranges See also: http://www.handprint.com/LS/ANC/stones.html H. Sapiens Cro Magnon H. heidelbergensis http://www.handprint.com/LS/ANC/evol.html#chart H. neanderthalensis H. erectus H. ergaster http://www.sciencemag.org/content/vol328/issue5979/images/me dium/328_680_F1.gif http://www.visionmagazine.com/archives/0712/images/culture_b elief.jpg Porgaigas New Guinea Still practice cannibalis G/wi Hunter gatherers Kalahari, Africa Amerind Caucasian (from Caucasus Mts Northern European Asian Human geographic races • • • • • • • • • Are races legacies of ancient H. erectus lineages? Did races originate after the rise of modern H. sapiens? Age of alleles Geographic origin of alleles Age of neutral alleles 4Ne generations from origin to fixation (± large error) 4 * 10,000 * 20 = 800,000 years H.erectus left Africa about 1,000,000 years ago Molecular methods can identify recent origin, but are at the limit of resolution for “proving” an older origin of races Gene genealogies Most recent common ancestor old medium young old Ages of alleles Time Fig. 1 Samples and sites from which DNA was retrieved R. E. Green et al., Science 328, 710-722 (2010) Published by AAAS Neandertal mtDNA Tree and Pairwise Divergence Green et al. 2008 Challenges for nuclear sequences: Ancestral polymorphism Most regions may be identical Contamination – 95%! Deamination: C->T Fig. 2 Nucleotide substitutions inferred to have occurred on the evolutionary lineages leading to the Neandertals, the human, and the chimpanzee genomes R. E. Green et al., Science 328, 710-722 (2010) Published by AAAS Fig. 4 Selective sweep screen R. E. Green et al., Science 328, 710-722 (2010) Published by AAAS Fig. 6 Four possible scenarios of genetic mixture involving Neandertals R. E. Green et al., Science 328, 710-722 (2010) Published by AAAS