* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Why haplotype analysis is not critical in genome wide association studies Derek Gordon
Genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genome evolution wikipedia , lookup
Genetic testing wikipedia , lookup
Gene expression programming wikipedia , lookup
Gene expression profiling wikipedia , lookup
Frameshift mutation wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Human genetic variation wikipedia , lookup
Point mutation wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Public health genomics wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Genome-wide association study wikipedia , lookup
Genome (book) wikipedia , lookup
SNP genotyping wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Genetic drift wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Dominance (genetics) wikipedia , lookup
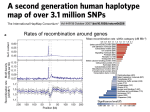
Why haplotype analysis is not critical in genome wide association studies Derek Gordon Department of Genetics Rutgers University Piscataway, NJ [email protected] Acknowledgements • Conference organizers and participants – Dr. Kui Zhang, Dr. David Allison, Dr. Hemant Tiwari, Dr. Jung-Ying Tzeng, Mr. Richard Sarver – Dr. Dan Schaid (The Pro) – Dr. Mike Province (The Provocateur) • Rutgers University – Dr. Steve Buyske • Rockefeller University – Dr. Jürg Ott Terminology Locus – Particular position, point, or place – Specific identifiable location on a chromosome Allele – Alternative forms of the same gene – Specific DNA sequence at a locus Polymorphic/polymorphism Genotype gggatc Allele 1 gggctc Allele 2 P M Locus 1 A or Genotype: 2 C – Specific alleles at each locus Haplotype – Specific alleles at many loci on the same chromosome Haplotype: 1 1 2 2 2 4 Terminology Locus – Particular position, point, or place – Specific identifiable location on a chromosome gggatc Allele 1 gggctc Allele 2 P M Locus Polymorphism consisting of single base pair change is called a Single Nucleotide Polymorphism (SNP). Question • What are reasons for not using haplotypes in genetic association analysis? Some answers • Curse of dimensionality – Temptation to simplify analysis. • Determining correct haplotype? • Biologically speaking, the SNP’s the thing! Do haplotypes provide statistical power gain over single marker tests for genetic association? NOT NECESSARILY! • Example -SNP that has two alleles, disease-causing (D) and wild-type (+). Frequencies in case and control populations are given in table at right. Allele Case Freq Control Freq D 0.1 0.05 + 0.9 0.95 Haplotype frequencies Haplotype Case Freq H1(containing 0.10 disease mutation) h2 (Middle p haplotype freq) h3 1 - p - 0.10 Control Freq 0.05 p + 0.025 1 - p - 0.075 Do haplotypes provide statistical power gain over single marker tests for genetic association? Statistical tests – Chi-square test of association on alleles (1 degree of freedom) or haplotypes (2 degrees of freedom). Compute minimum sample size for each test to detect association with 80% power at 10E-07 significance level. Efficiency = (Minimum Sample Size for Haplotype test)/(Minimum Sample Size for Allele Test). Middle Haplotype Frequency in Cases (p ) 0. 8 0. 84 0. 88 0. 64 0. 68 0. 72 0. 76 0. 6 0. 4 0. 44 0. 48 0. 52 0. 56 0. 2 0. 24 0. 28 0. 32 0. 36 0. 04 0. 08 0. 12 0. 16 0 Efficiency 1.2 1 0.8 0.6 0.4 0.2 0 Example – Alzheimer’s Disease One of the most well-documented and replicated results of a risk locus for late onset Alzheimer’s Disease (AD) is the APOE gene on Chromosome 19. There are three alleles at this locus, labeled ε2, ε3, and ε4. The last (ε4) is the risk allele for the AD. Fallin et al. (2001) Genome Res Vol. 11, Issue 1: 143-151 (Part of Table 3) Likelihood ratio test using haplotypes that flank (but do not include) SNPs in APOE gene is 45.64. Haplotype results less significant than single locus tests LRT value of 45.64 is smaller (and therefore, less significant) than value of 50.45 for SNP in APOE gene. Martin et al. (2000) : Am J Hum Genet. 67(2):383-94 (Figure 2). • Curse of dimensionality – Temptation to simplify analysis. • Determining correct haplotype? • Biologically speaking, the SNP’s the thing! Hierarchical clustering to reduce “curse of dimensionality” Example – Hoehe et al. (Hum Mol Genet. 2000 Nov 22;9(19):2895-908) estimated 52 haplotypes in African American cases and controls when testing for association of substance dependence with mu-opioid receptor gene. Hoehe et al. table of haplotypes Hierarchical clustering to reduce “curse of dimensionality” The large number of haplotypes made for difficult interpretation and for power loss. The authors proposed a hierarchical clustering method to reduce degrees of freedom by grouping similar haplotypes together. Clustering yielded a minimum p-value of 0.017 at 20th step (2 haplotype classes) Correction for correlated tests Levenstien et al. (BMC Bioinformatics. 2003 Dec 11;4:62) applied permutation methods to the Hoehe et al. data, replicating the clustering application in each permuted data set. The findings were that the minimum p-value less than or equal to 0.017 occurred in almost 70% of the permuted data sets. Correction for correlated tests In other words, the p-value of the minimum p-value was 0.70, indicating no significant association among haplotypes and substance abuse. This example points out risks of ad hoc (and invalid) statistical analyses that are necessarily developed to address the dimensionality problem with haplotypes. • Curse of dimensionality – Temptation to simplify analysis. • Determining correct haplotype? • Biologically speaking, the SNP’s the thing! Can we determine correct haplotypes for individuals? PLoS Genet. 2006 August; 2(8): e127. Results of haplotype-pair misclassification • For genes with substantial amount of recombination, some haplotype pairs had 100% misclassification rates for SNPHAP program and nearly 100% misclassification rates for PHASE program. • Curse of dimensionality – Temptation to simplify analysis. • Determining correct haplotype? • Biologically speaking, the SNP’s the thing! The SNP’s the thing! Genotypes are the more biologically relevant units of measurement for genetic association of genetic traits. Three base-pair code for determining Amino Acids Types of mutations www.ucl.ac.uk/~ucbhjow/b241/images/mutation.gif Types of mutations (deletion of single base pair) www.ucl.ac.uk/~ucbhjow/b241/images/mutation.gif If we type the SNP and only the SNP that causes the change in phenotype (e.g., missense, nonsense, or frameshift mutations), with sufficiently large sample size, we can determine the location of the mutation without haplotype analysis. Science 2005 Apr 15;308(5720):385-9. Only single locus SNP tests used to map AMD in Klein et al. study (Figure 1A). Nat Med (2008) Epub April 20. Authors sequenced GRK2 and GRK5 genes (no haplotyping) and found non-silent basepair change in GRK5 gene, resulting in two alleles: GRK5-Q41 (non-protective against heart failure) and GRK5-L41 (protective against heart failure). Survival curves for subjects with GRK5-Q41 allele using beta-blockers and subjects with GRK5-L41 allele not using beta-blockers (Figure 3c – Liggett et al). To summarize • Haplotype analysis increases the complexity of the analysis, in terms of the number of haplotypes introduced, the assumptions required, and the interpretation of the analysis. • Determination of correct haplotype is problematic. • Biological relevance of haplotypes? Finally… Will we even need haplotype analysis in the future? Can we always type the causative locus? In the near future, YES! Science (2008), Vol 311, pg 1544. Importance of typing causative SNP (PAWE-3D Website)