* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Coupling transcription, splicing and mRNA export
Cellular differentiation wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Transcription factor wikipedia , lookup
Cell nucleus wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
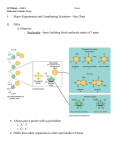
326 Coupling transcription, splicing and mRNA export Robin Reed Previous studies have led to the view that mRNAs are transported from the nucleus to the cytoplasm by machinery that is conserved from yeast to humans. Moreover, this machinery is coupled both physically and functionally to the pre-mRNA splicing machinery. During the past year, important new insights into the mechanisms behind this coupling have been made. In addition, recent advances have revealed mechanisms for the co-transcriptional loading of the export machinery on to mRNAs. Finally, a newly identified link between the nuclear exosome and the machineries required for transcription, 30 -end formation and mRNA export suggests that proper mRNP formation is co-transcriptionally monitored. Addresses Department of Cell Biology, 240 Longwood Avenue, Harvard Medical School, Boston, MA 02115, USA e-mail: [email protected] Current Opinion in Cell Biology 2003, 15:326–331 This review comes from a themed issue on Nucleus and gene expression Edited by Jeanne Lawrence and Gordon Hager 0955-0674/03/$ – see front matter ß 2003 Elsevier Science Ltd. All rights reserved. DOI 10.1016/S0955-0674(03)00048-6 Abbreviations ChIP chromatin immunoprecipitation EJC exon-junction complex NPC nuclear pore complex RNAi RNA interference RNP ribonucleoprotein TREX transcription export complex Introduction During expression of protein-coding genes, pre-mRNAs are transcribed in the nucleus and undergo several processing steps, including capping, splicing, 30 -end processing and polyadenylation. The mature mRNA is then exported through nuclear pore complexes (NPCs) to the cytoplasm for translation. Although distinct and highly complex cellular machines carry out each of these steps in the gene-expression process, growing evidence suggests the existence of gene-expression factories in which individual machines are functionally coupled. This coupling is obviously not obligatory, as virtually all of the coupled steps in gene expression can be uncoupled in various assays. Thus, coupling probably exists to enable the proofreading and streamlining of the entire process of gene expression in vivo. In this review, the discussion will focus on advances made in the past year in elucidating the Current Opinion in Cell Biology 2003, 15:326–331 mechanisms coupling splicing and transcription to mRNA export. In addition, new studies that link the nuclear exosome to the machineries involved in transcription, 30 end formation and mRNA export will be discussed. Readers are referred to several recent reviews for more extensive descriptions of coupling between the different steps in gene expression [1–7]. Coupling splicing to mRNA export The notion that splicing is linked to mRNA export first came from studies in metazoans showing that spliced mRNAs are assembled into a distinct ‘spliced mRNP’ complex that targets the mRNA for export (for a review, see [3]). This targeting involves the splicing-dependent recruitment of the mRNA export factor Aly via its direct interactions with the spliceosomal protein UAP56. In Saccharomyces cerevisiae, the counterparts of Aly and UAP56 also interact with each other directly and are required for mRNA export. In both S. cerevisiae and metazoans, Aly also interacts directly with Tap, which, together with its binding partner p15, associates with the NPC and is thought to be a general mRNA export receptor. These and other studies indicate that there is a conserved machinery for mRNA export, as shown in Figure 1 (for reviews, see [3,6,8,9]). In metazoans, where most genes contain introns, this conserved machinery is intimately tied to the splicing machinery. By contrast, in S. cerevisiae, where few genes contain introns, it appears that mRNA export is coupled to other steps in gene expression. Work initiated by the Moore group has shown over the past few years that in metazoans the conserved mRNA export machinery (UAP/Aly/Tap) and other components of the spliced mRNP are specifically recruited to a position 20 nucleotides upstream of exon–exon junctions ([10] and references therein). In addition to export proteins, this exon-junction complex (EJC) contains several proteins involved in nonsense-mediated decay (the degradation of mRNAs containing premature stop codons) and in the cytoplasmic localization of mRNAs. Thus, the EJC is thought to play multiple roles in mRNA metabolism. An intriguing feature of the EJC is that it is positioned on the mRNA at a specific site, but in a sequence-independent manner. Recently, Moore and colleagues have begun to determine the mechanism for positioning the EJC [10]. Their analysis revealed that numerous proteins (nine were detected) interact with the 50 exon in the catalytically active spliceosome. During exon ligation, a major restructuring of this region occurs, placing three other proteins (p170, p95 and p57) at the site where the EJC forms [10]. Aly and SRp20 (a member of www.current-opinion.com Coupling transcription, splicing and mRNA export Reed 327 Figure 1 Mattaj and co-workers [13] addressed the role of splicing in mRNA export by constructing a hybrid U1 snRNA in which exons, introns or random sequences were inserted. The authors concluded that mRNAs contain an ‘identity element’, which they defined as an unstructured sequence at least 300 nucleotides long. This identity element is sufficient to re-direct the hybrid RNA away from the U1 snRNP export pathway and into the UAP/Aly/Tap mRNA export pathway, even in the absence of splicing. Consistent with previous studies (for a review, see [3]), the implication of the new work is that splicing is not essential for export but increases the efficiency and/or fidelity of the process by promoting the more efficient recruitment of export factors. Spliceosome UAP ALY UAP Spliced mRNP ALY TAP Nucleus Cytoplasm Current Opinion in Cell Biology Conserved mRNA export machinery coupled to splicing. UAP56 (Sub2 in yeast), which is present in the spliceosome, recruits Aly (Yra1 in yeast) to the mRNA during splicing. UAP56 is replaced by the mRNA export receptor TAP/p15 (Mex67/MTR2 in yeast). the SR protein family of splicing factors) also contact this region in the EJC [10]. These five RNA-contacting proteins are therefore likely to play key roles in EJC formation. Once formed, the EJC is associated with mRNAs that are bound in the nucleus by the cap-binding protein CBP80 but not with mRNAs bound by the cytoplasmic eIF4E cap-binding protein [11]. The EJC proteins associate with CBP80-bound mRNAs both in the nucleus and in the cytoplasm, suggesting that the entire EJC is exported and then dissociates from the mRNA in the cytoplasm [11]. In yeast, Lei and Silver [12] used the chromatin immunoprecipitation (ChIP) method to show that Yra1 (yeast Aly) and Sub2 (yeast UAP56) associate preferentially with the intron region, in a splicing-dependent manner. Thus, it is possible that both splicingdependent recruitment of the mRNA export machinery and EJC formation are also conserved. www.current-opinion.com In an unexpected twist to the emerging story of the conserved mRNA export pathway and the EJC, a new Drosophila RNA interference (RNAi) study by Gatfield and Izaurralde [14] concludes that, although both Tap and UAP56 are required for mRNA export, Aly and the other EJC components are dispensable. The authors propose that an as-yet undiscovered adaptor protein(s) must exist that mediates the interaction between Tap and the mRNA. However, there are alternative possibilities; the RNAi-mediated knockdown of Aly and the other EJC proteins may not have been sufficient to eliminate their functions, or there may be a redundancy in factors. Indeed, the new conclusion is difficult to reconcile with previous work. For example, antibodies to Aly block mRNA export, and recombinant Aly stimulates mRNA export (for a review, see [3]). Moreover, the physical and functional conservation of UAP, Aly, and Tap from yeast to humans implicates these proteins as key players in mRNA export [3,8]. Coupling transcription to mRNA export Although the UAP/Aly/Tap pathway is conserved from yeast to humans, the export machinery is not solely recruited by a splicing-dependent mechanism. Indeed, 95% of S. cerevisiae genes as well as several metazoan genes lack introns. Nevertheless, in both metazoans and yeast, the conserved UAP/Aly/Tap pathway is required for the export of mRNAs derived from genes lacking introns ([14] and references therein). Thus, a central focus of recent work has been to determine the mechanisms for splicing-independent recruitment of this machinery. In earlier work, the Silver group shed light on this problem by providing evidence that the mRNA export machinery is co-transcriptionally recruited in S. cerevisiae (for a review, see [6]). During the past year, new advances have been made that support this view and that identify possible mechanisms for the recruitment. In one recent study in S. cerevisiae, Strasser et al. [15] showed that Sub2 and Yra1 specifically associate with the multisubunit THO complex, which is thought to be involved in transcription elongation (for a review, see Current Opinion in Cell Biology 2003, 15:326–331 328 Nucleus and gene expression [16]). Null mutants in any of the non-essential THO complex subunits (Hpr1, Tho2, Mtf1 and Thp2) also display an unusually high level of transcription-dependent recombination ([16,17] and references therein). This hyper-recombination phenotype may be a secondary consequence of the polymerase elongation defect, which exposes transcription units to recombination [16]. In the study by Strasser et al. [15], all four subunits of the THO complex were found to interact genetically and physically with bothYra1 and Sub2. In addition, the THO-complex null mutants are defective in mRNA export. Finally, ChIP assays revealed that THO complex components, as well as Yra1 and Sub2, associate with actively transcribed genes [15,18]. The THO complex together with the mRNA export proteins is designated the transcription export complex (TREX) [15]. Together, the data support a model in which the TREX complex plays a role in coupling transcription to mRNA export. Stutz and co-workers [18] have advanced this model further by showing that the THO/TREX component Hpr1 interacts directly with Sub2 and plays a role in recruiting Sub2 and Yra1 to actively transcribed genes. In another study in S. cerevisiae, Hurt and co-workers [19] identified what are likely to be new components of the conserved mRNA export machinery, with links to the transcription machinery. Performing a synthetic lethal screen with Yra1 (yeast Aly), they identified the Sac3 gene. The Sac3 protein was also identified by mass spectrometry by virtue of its association with Sub2 (yeast UAP56) in a pull-down assay from yeast lysates [15]. Sac3 was also shown to interact directly with Mex67 (yeast Tap). Thus, Sac3 interacts physically and functionally with all of the components of the UAP/Aly/TAP pathway. But the story does not end there; Sac3 also forms a complex with Thp1, a protein thought to function in transcription elongation, and Hurt and coworkers [19] showed that both Sac3 and Thp1 are required for mRNA export. Moreover, Sac3 also interacts with two nucleoporins (Nup 60 and Nup 1) that are located on the nucleocytoplasmic face of the NPC and that are required for mRNA export. On the basis of these observations, Hurt and coworkers proposed a model in which Sac3 first associates with the mRNP early in its formation, perhaps during transcription, and then, by interacting with both Mex67 and the two nucleoporins, facilitates docking of the mRNP at the nuclear-pore complex. Silver’s group [20] also identified Sac3 in a synthetic lethal screen using a known mRNA export factor, in this case MTR2 (the small subunit of the Mex67/MTR2 mRNA export receptor [Tap/p15 in metazoans]). Sac3 was shown to function in mRNA export, to interact genetically with several mRNA export factors and to associate physically with Mex67, MTR2 and specific nuclear-pore-complex-associated proteins involved in mRNA export. Immunolocalization of Sac3 revealed Current Opinion in Cell Biology 2003, 15:326–331 that it associates with the cytoplasmic fibrils of the NPC. Significantly, when Sac3 is mutated, Mex67 accumulates at the nuclear rim [20]. On the basis of these results, Silver and co-workers [20] propose that Sac3 plays a role in a terminal step in the process whereby Mex67 is translocated across the NPC. Considering both the Hurt and Silver studies, it is possible that Sac3 associates with the mRNA during transcription, plays a role in docking Mex67 to the nuclear pore and then functions in the translocation of Mex67 across the nuclear pore. Metazoan counterparts of Sac3 and Thp1 have been identified. Thus, it would not be surprising if these proteins play a role in mRNA export in metazoans. Links between mRNA export and other steps in gene expression Surprising new studies by the Jensen group [21] and the Stutz group [18] have linked the TREX complex to the nuclear exosome, a large complex of 30 to 50 exonucleases involved in numerous RNA processing and degradation pathways. The two new studies have shown genetic interactions between exosome and TREX complex components. Moreover, Yra1 interacts physically with the exosomal protein Rrp45 [18]. Previous studies have shown that TREX complex mutants generate transcripts that are truncated at their 30 ends (for a review, see [16]). Jensen and colleagues have now shown that in these mutants, as well as in mutants that affect proper 30 end formation, mRNAs are rapidly sequestered near their sites of transcription ([21] and references therein). Both this phenotype and the 30 -end truncation phenotype can be rescued by deletion of the exosome component Rrp6 [21]. Together, these studies led to the view that proper mRNP formation is monitored by the exosome, resulting in retention and destruction of aberrant mRNPs at their sites of transcription [7,18,21]. Significantly, another link was recently made between the exosome and transcription elongation factors (Spt5 and Spt6) [22]. In this study, Lis and co-workers showed that the exosome physically associates with Spt5 and Spt6 and is recruited to actively transcribed genes, possibly by these proteins. Together, the new data suggest a model for how the exosome, the TREX complex, Spt5 and Spt6 regulate transcription elongation, mRNP formation and export. In this model, Spt5 and Spt6 recruit the exosome to RNA polymerase II at an early stage of transcription (Figure 2). The TREX complex, which associates with the gene at a later stage of transcription, may physically protect the mRNP from the exosome or may even function as an exosome inhibitor (Figure 2a). When the TREX complex is missing (as in the TREX-null mutants), the 30 ! 50 exonuclease activity of the exosome destroys the growing mRNP, leading to the dissociation of RNA polymerase from DNA and thus blocking transcription elongation (Figure 2b). When both the TREX www.current-opinion.com Coupling transcription, splicing and mRNA export Reed 329 Figure 2 Exosome Spt5,6 RNAP II 5′ (a) TREX Exosome ALY Spt5,6 UAP RNAP II (b) 3′ Exosome S Spt5,6 RNAP II (c) Spt5,6 RNAP II 5′ ALY UAP Exported 3′ truncated/degraded Full length Not exported Not exported Current Opinion in Cell Biology In this model, coupling the transcription and export machineries to the exosome enables the monitoring of proper mRNP formation. The exosome is recruited during transcription, possibly by the elongation factors Spt5 and Spt6. When the TREX complex is present, a proper mRNP is made containing export proteins (Aly and UAP56) and the mRNA is exported (a). In the absence of the TREX complex, the exosome degrades the mRNA from the 30 to the 50 end and the transcription machinery dissociates (b). When both the TREX complex and the exosome are absent, transcription can occur but the mRNP lacks export proteins and is not exported (c). complex and the exosome are missing, full-length transcripts would be generated but not exported because the export machinery cannot be recruited (Figure 2c). Thus, the purpose of coupling the exosome to the TREX complex is to ensure that any transcripts that lack the TREX complex, and hence the mRNA export machinery, are targeted for destruction. Although ample evidence indicates that the mRNA export machinery is co-transcriptionally loaded onto mRNAs in yeast, 30 -end formation may play a more critical role in this loading. For example, Lei and Silver [12] find that Sub2 is not required for the recruitment of Yra1 to genes that lack introns. However, proper 30 -end formation is required to recruit this factor, whether or not an intron is present. This observation supports earlier studies indicating that 30 -end formation is important for mRNA export (for reviews, see [6,7]). In further support of this view, Rosbash’s group has recently reported that transcripts generated by T7 polymerase (i.e. not by RNA polymerase II) are exported only if 30 -end formation occurs normally [23]. This story has also been extended www.current-opinion.com by Hammel et al. [24]. In a screen to identify genes required for mRNA export, several factors involved in termination and 30 processing were obtained. The study also showed that the deletion of sequences in the mRNA that are involved in coupling termination and 30 end formation leads to the synthesis of transcripts that cannot be exported. Thus, the authors propose that termination, 30 processing and mRNA export are all coupled. Coupling transcription and splicing to mRNA export in metazoans Numerous recent studies in metazoans have provided compelling new evidence that transcription can affect splicing and vice versa (see for example [25–28]). Because of space restrictions, the discussion here will be limited to recent advances concerning the coupling of both transcription and splicing to mRNA export. In particular, Strasser et al. [15] identified the likely counterpart of the TREX complex in mammals. As in yeast, this complex contains mRNA export proteins (Aly and UAP56) as well as THO complex components (hTho2 and hHpr1). It is not yet known whether this putative mammalian Current Opinion in Cell Biology 2003, 15:326–331 330 Nucleus and gene expression Acknowledgements Figure 3 I am grateful to Ming-Juan Luo, Ed Hurt and Tom Maniatis for helpful comments on the review and to Claudine Christoforides for assistance in preparation of the manuscript. I apologize for references that were not cited because of space limitations. References and recommended reading ALY UAP Spliceosome Papers of particular interest, published within the annual period of review, have been highlighted as: of special interest of outstanding interest ALY UAP 1. Orphanides G, Reinberg D: A unified theory of gene expression. Cell 2002, 108:439-451. 2. Bentley D: The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol 2002, 14:336-342. 3. Reed R, Hurt E: A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 2002, 108:523-531. 4. Maniatis T, Reed R: An extensive network of coupling among gene expression machines. Nature 2002, 416:499-506. 5. Proudfoot NJ, Furger A, Dye MJ: Integrating mRNA processing with transcription. Cell 2002, 108:501-512. 6. Lei EP, Silver PA: Protein and RNA export from the nucleus. Dev Cell 2002, 2:261-272. 7. Jensen TH, Rosbash M: Co-transcriptional monitoring of mRNP formation. Nat Struct Biol 2003, 10:10-12. 8. Izaurralde E: A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol 2002, 81:577-584. 9. Dreyfuss G, Kim VN, Kataoka N: Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 2002, 3:195-205. TREX RNAP II Current Opinion in Cell Biology In this model, coupling transcription, splicing and export via the TREX complex targets the export machinery to exons. Metazoan introns are typically very large whereas the exons are minute. The TREX complex may associate with the spliceosome to allow co-transcriptional loading of the export machinery (Aly and UAP56) onto exons, which are destined for export, and to exclude this machinery from the introns, which are retained in the nucleus. TREX complex associates with the nascent pre-mRNA during transcription. Surprisingly, however, all of the mammalian TREX complex components were found in purified spliceosomes [29–32]. Thus, the TREX complex may function to link both transcription and splicing to mRNA export in metazoans. One model that may explain why the TREX complex is linked to splicing in metazoans concerns the structure of their genes. In metazoans, the abundant introns are typically thousands to tens of thousands of nucleotides in length and can be as large as several hundred thousand nucleotides. The exons, averaging only 100 nucleotides, are minute by comparison. Thus, as shown in Figure 3, the TREX complex may be associated with the spliceosome as a mechanism for cotranscriptionally targeting the mRNA export factors only to exons, which are destined for export, and not to the numerous enormous introns, which are retained and degraded in the nucleus. Conclusions Recent studies strengthen the view that mRNA export is coupled to other steps in gene expression including splicing, transcription, and 30 -end formation. In addition, new links between mRNA export, transcription and the nuclear exosome were revealed. Future directions will include a detailed characterization of the spliced mRNP and EJC that couples splicing to mRNA export. In addition, the role of the TREX complex in export and transcription, and the relationship between these processes and the nuclear exosome remain to be determined. Finally, the molecular basis for coupling 30 -end formation to mRNA export must be identified. Current Opinion in Cell Biology 2003, 15:326–331 10. Reichert VL, Le Hir H, Jurica MS, Moore MJ: 50 exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev 2002, 16:2778-2791. The authors explored formation of the EJC. The data show that a set of proteins contacts the exon during splicing and that extensive remodeling then occurs in this region as the exon junction is formed. 11. Lejeune F, Ishigaki Y, Li X, Maquat LE: The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 2002, 21:3536-3545. 12. Lei EP, Silver PA: Intron status and 30 -end formation control cotranscriptional export of mRNA. Genes Dev 2002, 16:2761-2766. 13. Ohno M, Segref A, Kuersten S, Mattaj IW: Identity elements used in export of mRNAs. Mol Cell 2002, 9:659-671. 14. Gatfield D, Izaurralde E: REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 2002, 159:579-588. 15. Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R et al.: TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417:304-308. A new complex containing mRNA export factors and transcription factors is identified in yeast and mammals. The complex may function in the co-transcriptional loading of mRNA export factors. 16. Jimeno S, Rondon AG, Luna R, Aguilera A: The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 2002, 21:3526-3535. 17. Aguilera A: The connection between transcription and genomic instability. EMBO J 2002, 21:195-201. 18. Zenklusen D, Vinciguerra P, Wyss JC, Stutz F: Stable mRNP formation and export require cotranscriptional recruitment of www.current-opinion.com Coupling transcription, splicing and mRNA export Reed 331 the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 2002, 22:8241-8253. This study shows that the THO/TREX complex component Hpr1 interacts with the mRNA export factor Sub2 and plays a role in recruiting Sub2 to the mRNA during transcription. The study also shows links between the TREX complex and the nuclear exosome, suggesting that proper mRNP formation is co-transcriptionally monitored. 19. Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E: The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 2002, 21:5843-5852. Sac3 is identified as a new mRNA export factor. Sac3 was shown to interact with specific proteins, leading to the proposal that Sac3 is loaded onto mRNA co-transcriptionally and later functions to dock the mRNP at the nuclear pore. 20. Lei EP, Stern CA, Fahrenkrog B, Krebber H, Moy TI, Aebi U, Silver PA: Sac3 is an mRNA export factor that localizes to the cytoplasmic fibrils of the nuclear pore complex. Mol Biol Cell 2002, 14:836-847. This study also identifies Sac3 as a new mRNA export factor. The authors show that Sac3 localizes to the cytoplasmic fibrils and may play a role in a terminal step of mRNP translocation. 21. Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH: Interactions between mRNA export commitment, 30 -end quality control, and nuclear degradation. Mol Cell Biol 2002, 22:8254-8266. This study reports a link between the TREX complex and the nuclear exosome, providing further evidence for the author’s proposal that the exosome monitors proper mRNP formation co-transcriptionally. 22. Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT: The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 2002, 420:837-841. This study is the first to show that the nuclear exosome is recruited to actively transcribed genes. The elongation factors Spt5 and Spt6 are in a complex with the exosome and may play a role in recruiting it to active genes. The study further strengthens the notion that proper mRNP formation is monitored during transcription. www.current-opinion.com 23. Dower K, Rosbash M: T7 RNA polymerase-directed transcripts are processed in yeast and link 30 end formation to mRNA nuclear export. RNA 2002, 8:686-697. This study shows that 30 -end formation plays a key role in mRNA export in yeast. 24. Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN: Coupling of termination, 30 processing, and mRNA export. Mol Cell Biol 2002, 22:6441-6457. These authors report that termination and 30 end processing are coupled to mRNA export in yeast. 25. Kwek KY, Murphy S, Furger A, Thomas B, O’Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A: U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 2002, 9:800-805. 26. Auboeuf D, Honig A, Berget SM, O’Malley BW: Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 2002, 298:416-419. 27. Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ: Promoter proximal splice sites enhance transcription. Genes Dev 2002, 16:2792-2799. 28. Kadener S, Fededa JP, Rosbash M, Kornblihtt AR: Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci USA 2002, 99:8185-8190. 29. Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ: Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 2002, 8:426-439. 30. Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R: Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci USA 2002, 99:16719-16724. 31. Zhou Z, Licklider LJ, Gygi SP, Reed R: Comprehensive proteomic analysis of the human spliceosome. Nature 2002, 419:182-185. 32. Rappsilber J, Ryder U, Lamond AI, Mann M: Large-scale proteomic analysis of the human spliceosome. Genome Res 2002, 12:1231-1245. Current Opinion in Cell Biology 2003, 15:326–331