* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download contorl-of-cell-cycle 105 kb contorl-of-cell
Survey
Document related concepts
Extracellular matrix wikipedia , lookup
Spindle checkpoint wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Programmed cell death wikipedia , lookup
Signal transduction wikipedia , lookup
Cell culture wikipedia , lookup
Endomembrane system wikipedia , lookup
Cellular differentiation wikipedia , lookup
Cell nucleus wikipedia , lookup
Cytokinesis wikipedia , lookup
Cell growth wikipedia , lookup
Transcript
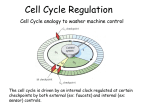
How did experiments with cell fusion, oocytes and yeast lead to our current understanding of the control of the cell cycle? (PLAN) Intro: Regulation of cycle is key to cell survival. Uses detection and repair of DNA damage and prevention of uncontrolled proliferation which would lead to tumour growth. Sequential, so errors carried forward if not repaired at checkpoints. Cell fusion: 4 stages: G1, s, G2, M G1= growth, high biosynthesis. s= DNA replication m=mitosis With cultured mammalian cells, one in s, one in G1 = 2s G1 cell enters s early shown by adding radioactive thymine, incorporated into new DNA G1/S/G2 + M cell= early M (nuclear envelope breaks down and chromatin condenses) Therefore, positive factors drive entry in s and m. G2+s doesn't drive G2 back to s, factors driving s can't diffuse into G2 nucleus, this highlights sequential nature of cycle. oocytes/early embryos: Frog oocytes: experiments with inhibitors showed protein synthesis required for increase in Mitosis promoting factor(MPF). Masui found frog oocytes arrested in G2 entered 2nd meiotic division after MPF (from mature embryos) injection. Tim Hunt: sea urchin embryo cells divide in synchrony. use radioactive amino acids and samples removed every 10 mins. Proteins extracted and amount analyzed by gel electrophoresis. one protein rose in interphase and early M phase and fell in anaphase =cyclin B cyclin B+cdk=mpf complex MPF activity rises/falls with cyclin B concentration. Cyclin B degradation= lower MPF activity = exit from M into G1, can't return to M until next cycle (more evidence for sequential nature). Yeast: Saccharomyces cerevisiae (budding yeast) and Schizosaccharomyces pombe (fission yeast)temperature sensitive mutants studied. Cell cycle paused at non-permissive temperatures. Microscopy observation experiments= Hartwell: S.cerevisiae, bud size increases during cycle, mutants identified at non-permissive temp = cdc mutants, arrested at G1. Nurse: S.pombe: cdc mutants elongate but don't divide. wee mutants divide before parental cell is ready to form short cells. Complementation experiments= haploid yeast mutants+human plasmid library (high conservation) to find which plasmids complement to allow mutants to undergo normal replication. S.cerevisiae: cdc28 gene, initiates S phase. S.pombe: cyclin dependent kinase (cdk1)= early entry into mitosis. With exp on cyclin b, MPF structure found. MPF phosphorylates proteins to initiate mitosis, eg nuclear envelope proteins to allow breakdown. wee1= cdk1 tyrosine 15 kinase, high activity in interphase. Activity drops near start of mitosis, tyrosine phosphatase dephosphorylates, cdk1 becomes active, M entry. Another checkpoint at the end of G1, into s or G0, experiments with S.cerevisiae: cdk4/6 regulation. Competitive inhibitor= cdkI p16 prevents cyclin D/cdk4/6 interaction. High cyclin D conc overcomes this, S initiated. conclusion: Experiments with cell fusion, oocytes, early embryos and yeast show sequential nature of cell cycle, highlighting the importance of regulatory steps and precise info on checkpoints (when, how, where, genes responsible). Applications in study of tumor formation (cancer), viral infection, mutations, DNA damage.