* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Etiology of hypoplastic left heart syndrome: insights from mutant
Survey
Document related concepts
Public health genomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Frameshift mutation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Birth defect wikipedia , lookup
Point mutation wikipedia , lookup
Fetal origins hypothesis wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Transcript
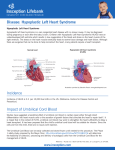
Etiology of hypoplastic left heart syndrome: insights from mutant mouse models Xiaoqin Liu1, Shazina Zaed1, Levin Chen1, George Gabriel1, Molly Schwartz1, Kevin Peterson2, Abha Bais1, Hisato Yagi1, Karen Bentley3, Nikolai Klena1, You Li1, Brian Gibbs1, William Devine4, Linda Leatherbury5, Maliha Zahid1, Madhavi Ganapathiraju6, Dennis Kostka1, Stephen Murray2, George Porter3, Cecilia W. Lo1 1 Department of Developmental Biology, 4Department of Pathology, 6Department of Biomedical Informatics, University of Pittsburgh School of Medicine, Pittsburgh, PA, 2The Jackson Laboratory, Bar Harbor Maine, 3Department of Pediatrics Division of Cardiology, University of Rochester School of Medicine, Rochester, NY, 5Children’s National Heart Institute, Children’s National Medical Center, Washington, DC. Hypoplastic left heart syndrome (HLHS), a congenital heart disease with high morbidity and mortality, is a left-sided structural heart disease characterized by severe hypoplasia of the left ventricle (LV), aorta and mitral valve. While a genetic etiology for HLHS is well supported by familial studies, the specific genes and mechanism of disease pathogenesis for HLHS is not well understood. In this study, we report findings from the first animal model of HLHS recovered from a large-scale recessive mouse mutagenesis screen. From over 100,000 fetuses ultrasound scanned, only 9 fetuses from 8 mutant lines were identified with HLHS, making it one of the rarest congenital heart disease phenotype recovered from the screen. Mutation recovery by whole exome sequencing analysis showed no mutations are shared in common among the 8 HLHS mutant lines. Detailed analysis of the Ohia HLHS mutant line identified mutations in two genes causing HLHS: Sap130, a protein involved in chromatin remodeling, and Pcdha9, a protocadherin mediating cell-cell adhesion. Using CRISPR/CAS gene-editing, we generated mouse embryos with Sap130 and Pcdha9 mutations that showed the same constellation of left-sided heart lesions seen in HLHS. Further analysis of Ohia HLHS mutants revealed defect in cell cycle progression with increased apoptosis in the left but not right ventricle, but no change was observed for cardiomyocte cell size. Ultrastructural analysis with electron microscopy and functional assays revealed defects in mitochondrial maturation and cardiomyocte differentiation. RNAseq transcriptome profiling showed striking gene expression alterations in the LV associated with pathways regulating energy metabolism, cell proliferation/cell death, cancer, chromatin remodeling, and Tgfβ signaling. Together, the findings from the mutant mouse models show HLHS is genetically heterogeneous with an oligogenic etiology. We demonstrated that the LV hypoplasia in the Ohia HLHS mutant is associated with cell intrinsic defects involving abnormalities in cell cycle regulation, mitochondrial maturation and cardiomyocyte differentiation. Supported by funding from NIH U01-HL098180