* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Essentials of Glycobiology Lecture 6 (7) April 7th. (9) 1998 Ajit Varki
Cell growth wikipedia , lookup
Cell culture wikipedia , lookup
Protein moonlighting wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Cellular differentiation wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell membrane wikipedia , lookup
Cytokinesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Signal transduction wikipedia , lookup
Endomembrane system wikipedia , lookup
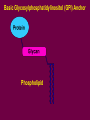
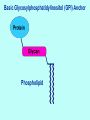
Essentials of Glycobiology Lecture 8 April 8, 2004 Hud Freeze Structure, biosynthesis and general biology of Glycophospholipid (GPI) Anchors Major Glycan Classes in Animal Cells CHONDROITIN SULFATE HYALURONAN P GLYCOSAMINOGLYCANS HEPARAN SULFATE S S S S S NS NS -O-Ser Proteoglycan N-LINKED CHAINS Ac O-LINKED CHAIN GLYCOPHOSPHOLIPID ANCHOR P S O Ser/Thr GLYCOSPHINGOLIPID N Asn N Asn INOSITOL Glycoprotein Ac Sialic Acids INSIDE O Ser Etn P NH 2 OUTSIDE O-LINKED GlcNAc S S Ser-O- P S Basic Glycosylphosphatidylinositol (GPI) Anchor Phospholipid Lecture Overview Historical Background Defining the Core Structure Biosynthesis & Transfer of GPI Anchors The Signal for Addition of GPI Anchors Occurrence and Variations in Nature Postulated Biological Roles Genetic Disorders Perspectives & Future Directions Discovery of GPI-Anchors. 1963-First data suggests protein-lipid anchors: crude bacterial phospholipase C releases alkaline phosphatase from mammalian cells. mid-1970’s Hiro Ikezawa in Japan, and Martin Low in the U.S. show that purified bacterial phosphatidylinositol phospholipase C releases some enzymes, e.g.,alkaline phosphatase, from cell surfaces. Propose Inositolcontaining phospholipid protein linkage Alan Williams in U.K. notes that antigen Thy-1 properties of glycolipid and glycoprotein. However: No structural data! GPI-anchors? Really? Discovery of GPI-Anchors The C-terminus of Thy-1 glycoprotein found to have both fatty acids and ethanolamine. In 1981, Tony Holder and George Cross groups showed that soluble form of the variant surface glycoprotein (sVSG) of African trypanosomes contains an immunocrossreactive carbohydrate (CRD) attached to its Cterminus via an amide linkage involving ethanolamine. Mervyn Turner’s group showed that trypanosomes contain an enzyme which rapidly releases the membraneassociated VSG (mfVSG) upon cellular damage. mfVSG becomes water soluble. sVSG so rapid membrane form is only detected by rapidly boiling trypanosomes in (SDS) prior to electrophoresis. Discovery of GPI-Anchors. 1985: Hart & Englund groups at Johns Hopkins show that the lipid-anchor on VSG is added within one minute of the polypeptide’s synthesis in the endoplasmic reticulum (ER). They postulate a pre-assembled membrane anchor is attached en bloc. 1985: Michael Ferguson and colleagues at Oxford publish a tour de force structural analysis of the glycolipid attached to the mfVSG of trypanosomes. These studies structurally define the term ‘glycosylphosphatidylinositol’ (GPI). THE LESSON: SHOW ME THE STRUCTURE!!! Basic Glycosylphosphatidylinositol (GPI) Anchor Phospholipid Examples of GPI-Anchored Proteins Cell surface hydrolases alkaline phosphatase acetylcholinesterase 5’ nucleotidase Adhesion molecules neural cell adhesion molecule heparan sulfate proteoglycan Protozoal antigens trypanosome VSG leishmanial protease plasmodium antigens Mammalian antigens carcinoembryonic antigen Thy-1 Others scrapie prion protein folate receptor decay accelerating factor Structure of the Basic GPI Anchor GPI-Linked Protein = Mannose (Man) NH 2 = Glucosamine Etn P = Ethanolamine Etn P = Phosphate PNH Defect NH 2 Pig-A INOSITOL P Cell Surface Membrane PLANTS ALSO MAKE GPI-ANCHORS QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Casper Vroemen,http://www.dpw.wau.nl/molbi/mediacenter/images/embryo11.jpg Studying GPI Biosynthesis in vitro thin layer chromatography F 30 °C O cell membranes salts, buffers radiolabeled sugardonor add solvents spin evaporate O F Structural Analysis of the GPI Anchor Enzymatic and chemical cleavage sites are useful in identifying GPI anchored membrane proteins Proposed branched pathway for biosynthesis of mammalian GPI anchors Examples of C-Terminal Sequences Signaling the Addition of GPI-Anchors Protein GPI-Signal Sequence Acetylcholinesterase (Torpedo) NQFLPKLLNATAC DGELSSSGTSSSKGIIFYVLFS ILYLIFY Alkaline Phosphatase (placenta) TACDLAPPAGTTD AAHPGRSVVPALLPLLAGTLLLLETATAP Decay Accelerating Factor HETTPNKGSGTTS GTTRLLSGHTCFTLTGLLGTLVTMGLLT PARP (T. Brucei) EPEPEPEPEPEP G AATLKSVALPFAIAAAALVAAF Prion Protein (hamster) QKESQAYYDGRRS SAVLFSSPP VILLISFLIFLMVG Thy-1 (rat) Variant Surface Glycoprotein (T. Brucei) KTINVIRDKLVKC GGISLLVQNTSWLLLLLLSLSFLQATDFISI ESNCKWENNACKD SSILVTKKFALTVVSAAFVALLF Bold AA is site of GPI attachment Sequence to right is cleaved by the transpeptidase upon Anchor addition Rules for C-Terminal Sequences Signaling the Addition of GPI-Anchors Residue to which anchor is attached (termed w site) and residue two amino acids on carboxyl side (w + 2 site) always have small side-chains w + 1 site can have large side-chains. w + 2 site followed by 5 to 10 hydrophilic amino acids, Next, add fifteen to twenty hydrophobic amino acids at or near the carboxy-terminus GPI Anchor Functions Dense packing of Proteins on Cell Surface Increased Protein mobility on Cell Surface Targeting of proteins to Apical Domains Specific release from Cell Surface Control of Exit from ER? Developmental regulation of protein expression? Generation of Protein Complexity Signal transduction? Toxin Binding Parasite Cell structure Possible Role of the GPI-Anchor in ER Exit Negative Signal: Retention mechanism is displaced by anchor transport vesicles - - Positive Signal: Anchor is recognized by packaging machinery transport vesicles + + UPS AND DOWNS OF GPI-LINKED PLACENTAL ALKALINE PHOSPHATASE QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. Frances J. Sharom www.chembio.uoguelph.ca/ sharom/ Sean Munro, Cell, 115, 377-388, Nov 2003 QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. CHONDROITIN SULFATE HYALURONAN P GLYCOSAMINOGLYCANS S S Ser-O- HEPARAN SULFATE Paroxysmal Nocturnal Hemoglobinuria: Somatic Loss of Glycophospholipid Anchors in Hematopoietic Stem Cells N-LINKED CHAINS S S S S S NS NS Ac O-LINKED CHAIN GLYCOPHOSPHOLIPID ANCHOR P S O Ser/Thr GLYCOSPHINGOLIPID -O-Ser N Asn N Asn NH 2 INOSITOL Glycoprotein Ac OUTSIDE Sialic Acids INSIDE O-LINKED GlcNAc O Ser Etn P P S Taroh Kinoshita QuickTime™ and a TIFF (Uncompressed) decompressor are needed to see this picture. • The first step in biosynthesis of the GPI anchor requires at least four genes • One of them, PIG-A is an X-linked gene Mutation in PNH MUTATIONS IN DOL-P-MAN SYN AND USE Biosynthesis of GPI anchors Paroxysmal Nocturnal Hemoglobinuria An acquired clonal hematopoietic stem cell disorder characterized by intravascular hemolytic anemia. Abnormal blood cells lack GPI-anchored proteins due to a mutation in the PIG-A gene. Lack of GPI-anchored complement regulatory proteins, such as decay-accelerating factor (DAF) and CD59, results in complement-mediated hemolysis and hemoglobinuria. Factors that determine why mutant clones expand have not been determined. Paroxysmal Nocturnal Hemoglobinuria Pre existing PNH clones have a conditional growth advantage depending on some factor present in the marrow environment of PNH patients. However, cells with the PNH phenotype have been found at a frequency of 22 per million in normal individuals. These rare cells were collected by flow sorting and had PIG-A mutations. Thus, PIG-A gene mutations are not sufficient for the development of clinically evident PNH. NON-POLITICAL FLIP-FLOP: A GOOD THING Quic kTime™ and a TIFF (Uncompress ed) dec ompres sor are needed to s ee this pic ture. Dol Quic kTime™ and a TIFF (Uncompress ed) dec ompres sor are needed to s ee this pic ture. CDG-If PTPATIENT WITH DPM1 DEFICIENCY Decreased Dol-P-Man synthesis PATIENT HAS MPDU1 DEFICIENCY Inefficient use of Dol-P-Man and Dol-P-Glc Both patients are blind with severe developmental delay Pathology may result from impaired N-linked or GPI-anchor synthesis FUTURE PERSPECTIVES THE FUNCTION OF GPI-ANCHORS IS STILL UNRESOLVED LIKE THE FUNCTIONS OF GLYCOYSLATION MAYBE ALL THE THEORIES ARE CORRECT