* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download InsP 3 R domains - Yale School of Medicine
Survey
Document related concepts
Mechanosensitive channels wikipedia , lookup
Cell encapsulation wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Purinergic signalling wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
Cellular differentiation wikipedia , lookup
Endomembrane system wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
P-type ATPase wikipedia , lookup
List of types of proteins wikipedia , lookup
Transcript
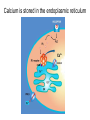
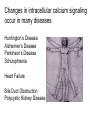
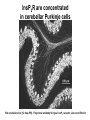
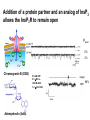
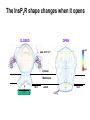
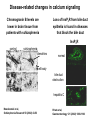
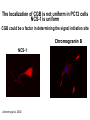
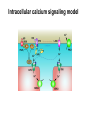
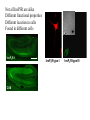
What the cryoEM structure of the InsP3 receptor can tell us about intracellular calcium signaling 25Å Barbara Ehrlich Pharmacology June 10, 2005 Calcium Calcium is stored in the endoplasmic reticulum CGB Changes in intracellular calcium signaling occur in many diseases Huntington’s Disease Alzheimer’s Disease Parkinson’s Disease Schizophrenia Heart Failure Bile Duct Obstruction Polycystic Kidney Disease InsP3R are concentrated in cerebellar Purkinje cells Rat cerebellar slice [12 days PN] . Polyclonal antibody for type I InsP3 receptor, Llano and Ehrlich InsP3R domains Ligand-Binding Domain N Regulatoy Domain C Channel Domain Patel S, Joseph SK, Thomas AP. Cell Calcium 1999 Mar;25(3):247-64 The InsP3R is a large protein that makes an ion channel kDa 250 100 50 Popen 0% 2% Addition of a protein partner and an analog of InsP3 allows the InsP3R to remain open Popen 0% 2% Chromogranin B (CGB) Adenophostin (AdA) 0.5 mM ATP 0.5 mM Ca2+ 100 nM AdA 1.0 mg/ml CGB 96% Examples of particles used for 3D reconstruction Match between model projections and class averages q, y q, y 0.0, 0.0 4.8, 0.0 33.6, 20.0 48.0, 7.5 57.6, 38.6 57.6, 45.0 67.2, 54.0 76.8, 45.0 86.4, 33.8 86.4, 39.4 CryoEM structure of the InsP3R using two analysis protocols InsP3R Closed State 24 A resolution Jiang et al. (2002) Current Closed State The InsP3R in the closed and open state closed open The InsP3 binding domain fits on the “arms” 25Å CLOSED Low high Cytosol Membrane 50 Å Lumen The InsP3R shape changes when it opens CLOSED OPEN Low AdA, ATP, Ca2+ high Cytosol Membrane 50 Å CGB4 50 Å Lumen CGB4 What we have learned already Structure changes during activity surprisingly large conformational changes suggests there is a mechanical gate Good idea where InsP3 binds the shape of the binding pocket will suggest classes of compounds to test as agonists and antagonists Disease-related changes in calcium signaling Chromogranin B levels are lower in brain tissue from patients with schizophrenia Loss of InsP3R from bile duct epithelia is found in diseases that block the bile duct InsP3R control schizophrenia dendrites normal cell body bile duct obstruction hepatitis C Nowakowski et al, Schizophrenia Research 58 (2002) 43-53 Hirata et al, Gastroenterology 121 (2002) 1088-1100 What we hope to learn Can we design new channel openers and closers? Where do the regulatory factors bind? Can these regulatory proteins be used as pharmacological agents? Can the structure tell us why the InsP3Rs cluster and go to distinct places in the cell? 50 Å CGB4 Collaborators Juliana Rengifo Bill Grenawitzke Chi-un Choe Nils Nicolay Qiu-Xing Jiang Fred Sigworth Edwin Thrower David Chester Brenda DeGray Mike Nathanson Daniel Hertle Mark Yeckel Comparison of the intracellular calcium release channels InsP3R type l RyR type 1 The localization of CGB is not uniform in PC12 cells NCS-1 is uniform CGB could be a factor in determining the signal initiation site Chromogranin B NCS-1 Johenning et al, 2002 Intracellular calcium signaling model The amplitude of the calcium transient can be modulated Dolmetsch, Xu and Lewis, 1998 The frequency of the calcium oscillation can be modulated Dolmetsch, Xu and Lewis, 1998 Not all InsP3R are alike Different functional properties Different location in cells Found in different cells CA3 InsP3 R 1 InsP3R type I CA1 InsP3R type III CGB CA3 CA1 CA3 CA1