* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 10: Control of gene expression
Koinophilia wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome (book) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Oncogenomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Genome editing wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
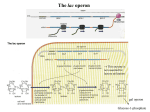
Lecture 10: Control of gene expression 1. One gene – one enzyme hypothesis 2. Lac operon 3. Negative and positive regulation Lac repressor bound to DNA The Nobel Prize in Physiology or Medicine 1958 "for their discovery that genes act by regulating definite chemical events" George Wells Beadle 1/4 of the prize California Institute of Technology (Caltech) Edward Lawrie Tatum 1/4 of the prize Rockefeller Institutute for Medical Research, NY "for his discoveries concerning genetic recombination and the organization of the genetic material of bacteria" Joshua Lederberg 1/2 of the prize University of Wisconsin Madison, WI Experiments of Beadle and Tatum, in 1940-ies, were the first to shed light on general mechanisms of the gene function. They studied mutations in the arginine biosynthetic pathway in Neurospora. Arginine has two related substances: citrulline and ornithine that are converted into arginine by the pathway that must involve several enzymes: Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine Search for mutants in amino acid biosynthesis Growth of some arg- mutants can be supplemented not only by arginine, but also by related compounds: Mutant arg 1 arg 2 arg 3 Ornithine + - Citrulline + + - Arginine + + + (plus means growth, and minus means no growth) The compound which is made the latest (Arg) supports growth of the largest number of mutants The compound which is made the earliest (Orn) supports growth of smallest number of mutants Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine how to explain the observed growth pattern? Mutant arg 1 arg 2 arg 3 Ornithine + - Citrulline + + - Arginine + + + - they hypothesized that each mutant is deficient in production of certain enzyme of the pathway, e.g. arg1 could not produce Enzyme X. Then this mutant cannot convert the precursor into ornithine. But given ornithine, it still can make citrulline and arginine. Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine arg1 Mutant arg 1 arg 2 arg 3 arg2+ Ornithine + - arg3+ Citrulline + + - Arginine + + + - mutant arg2 could not produce Enzyme Y. It can make ornithine (arg1+ allele) but cannot convert it into citrulline. That’s why its growth is not supported by ornithine. Citrulline, however, supports Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine arg1+ Mutant arg 1 arg 2 arg 3 arg3+ arg2 Ornithine + - Citrulline + + - Arginine + + + - mutant arg3 could not produce Enzyme Z. It can make ornithine and citrulline but cannot convert citrulline into arginine. That’s why its growth is not supported by ornithine or citrulline and is supported only by arginine. Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine arg1+ Mutant arg 1 arg 2 arg 3 arg2+ Ornithine + - arg3 Citrulline + + - Arginine + + + Enz. X Enz. Y Enz. Z Precursor -----> Ornithine ------> Citrulline -----> Arginine arg1+ arg2+ arg3+ ‘One gene – one enzyme’ hypothesis: genes control biosynthetic pathways by encoding enzymes that catalyze specific steps of the pathways Growth of E. coli with two energy sources: glucose and lactose Phase II: utilization of lactose Phase I: utilization of glucose ... nor is the metabolic chart Ability of E.coli cells to utilize lactose depends on production of the enzyme β-galactosidase Phenotype Minimal Medium with glucose Lac+(inducible) Lac- (negative) Lac++ (constitutive) - (no enzyme) + Minimal Medium with lactose + (enzyme is produced) + lac operon A group of genes that are regulated together and are closely linked to each other is called an operon Genes of the same operon are transcribed together as a polycistronic (= polygenic) mRNA Lac operon is located at 8 min of the E. coli chromosome map. It contains three genes lacZ, lacY and lacA. The respective three enzymes of the lac operon are induced simultaneously. lacP (promoter) and lacO (operator) are the operon’s regulatory regions lacZ- or lacY- mutations cause Lac- phenotype lac operon An additional class of mutations was found close to lac operon. These mutations caused constitutive phenotype (Lac++), and it was concluded that they affect ‘inducibility’ locus. Hence the name lacI. The lacI gene encodes lac repressor that inhibits the lac lacZ- or lacY- mutations cause Lac- phenotype operon. The operon is derepressed by lacI- mutations case Lac++ phenotype lactose in the medium. The Nobel Prize in Physiology or Medicine 1965 "for their discoveries concerning genetic control of enzyme and virus synthesis" François Jacob b. 1920 Jacques Monod 1910-1976 André Lwoff 1902-1994 lac repressor can reversibly attach to lac operon Lac+ phenotype lactose absent lactose present DNA –binding site Normal lac-repressor can interact with (bind to): 1) DNA (lac-operon) and 2) inducer (lactose) lactose –binding site Mutant forms of lac repressor: cannot attach to lac operon (lacI-) or cannot detach from it (lacIS) Lac++ phenotype lactose may be absent or present Lac- phenotype lactose may be absent or present Mutant lacIS repressor is dominant to normal lacI+ Meroploid F+ cell Lac- phenotype F’ lacI+ / lacIS lacZ+ (lacIS is in cis to lacZ+) F’ lacIS / lacI+ lacZ+ (lacIS is in trans to lacZ+) The same phenotype Lac- Normal lacI+ repressor is dominant to mutant lacIMeroploid F+ cell Lac+ phenotype Normal lacI+ repressor is dominant to mutant lacIMeroploid F+ cell Lac+ phenotype F’ lacI+ / lacI- lacZ+ (lacI+ is in trans to lacZ+) F’ lacI- / lacI+ lacZ+ (lacI+ is in cis to lacZ+) The same phenotype Lac+ Mutant lacOC operator in cis to lacZ is epistatic to all types of lac repressor Lac++ phenotype F’ lacI+ / lacOc lacZ+ F’ lacIS / lacOc lacZ+ F’ lacI- / lacOc lacZ+ Any type or configuration of lacI, provided that lacOC is in cis to lacZ+ the same phenotype Lac++ Mutant lacOC operator in trans to lacZ has no effect F’ lacOc / lacO+ lacZ+ Now the phenotype depends on the type of the repressor present: no epistatic effect of lacOC on alleles of lacI Carrot and whip on the molecular level cyclic AMP, a regulator of many biological processes the formulas are not for memorization glucose cAMP glucose Glucose is low cAMP transcription is activated