* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Differential Enzyme Targeting As an Evolutionary Adaptation to
Bisulfite sequencing wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Group selection wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Expanded genetic code wikipedia , lookup
Metagenomics wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Species distribution wikipedia , lookup
Genetic code wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
DNA barcoding wikipedia , lookup
Helitron (biology) wikipedia , lookup
Koinophilia wikipedia , lookup
Microevolution wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
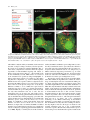
Differential Enzyme Targeting As an Evolutionary Adaptation to Herbivory in Carnivora Graeme M. Birdsey,* Jackie Lewin, Andrew A. Cunningham,à Michael W. Bruford,§ and Christopher J. Danpure* *Department of Biology, University College London, EM Unit, Royal Free and University College Medical School, àInstitute of Zoology, London, UK; §Cardiff School of Biosciences, Cardiff University, Cardiff, UK Not all members of the order Carnivora are carnivorous. Some are omnivorous, and a few, such as the giant panda, Ailuropoda melanoleuca, are almost exclusively herbivorous. Although a number of adaptations to increased plant-eating are recognized within Carnivora, few have been studied at the molecular level. One molecular adaptation to diet that is spread widely across Mammalia is the differential intracellular targeting of the intermediary metabolic enzyme alanine:glyoxylate aminotransferase (AGT), which tends to be mitochondrial in carnivores, peroxisomal in herbivores, and both mitochondrial and peroxisomal in omnivores. In the present study, we have analyzed the targeting of AGT in Carnivora in relation to species’ natural diets. We show not only that there has been an adaptive shift in AGT targeting from the mitochondrion toward the peroxisome as diets have shifted from being mainly carnivorous to ones that are more omnivorous and herbivorous but also that in one lineage, namely that of the giant panda, there is evidence for positive selection pressure at the molecular level on the AGT mitochondrial targeting sequence to decrease its efficiency, thereby allowing more AGT to be targeted to the peroxisomes. Introduction The subcellular distribution of the intermediary metabolic enzyme alanine:glyoxylate aminotransferase (AGT) has changed on numerous occasions during the evolution of mammals, apparently under the influence of dietary selection pressure. There is a tendency for AGT to be mitochondrial in carnivores (including insectivores), peroxisomal in herbivores, and both mitochondrial and peroxisomal in omnivores (Danpure et al. 1990,1994). AGT catalyses the detoxification of the intermediary metabolite glyoxylate to glycine, preventing it from being oxidized to oxalate. Too much oxalate can be life threatening because its calcium salt is poorly soluble and readily crystallizes out in the kidney and urinary tract, causing blockages and organ failure (Danpure and Smith 1996). For glyoxylate detoxification to be efficient, AGT must be concentrated at the site of glyoxylate synthesis. This is likely to be different in carnivores and herbivores because the main dietary precursor of glyoxylate in herbivores is thought to be glycolate, which is metabolized to glyoxylate in the peroxisomes (Noguchi 1987), whereas in carnivores it is more likely to be hydroxyproline that is converted to glyoxylate in the mitochondria (Takayama et al. 2003). The best evidence that the correct subcellular distribution of AGT is important in at least some species comes from the potentially lethal human hereditary kidney stone disease primary hyperoxaluria type 1 (PH1) (Danpure 2001). Although in most normal humans AGT is peroxisomal, in many PH1 patients AGT is mistargeted to the mitochondria (Danpure et al. 1989). Mistargeted AGT remains catalytically active but is unable to fulfill its metabolic role of glyoxylate detoxification efficiently. As a result, oxalate synthesis increases and calcium oxalate crystallizes out, usually as stones, in the kidney and urinary tract. Key words: molecular adaptation, alanine:glyoxylate aminotransferase, dietary selection pressure, protein targeting, mitochondria, peroxisomes. E-mail: [email protected]. Mol. Biol. Evol. 21(4):632–646. 2004 DOI: 10.1093/molbev/msh054 Advance Access publication January 22, 2004 Molecular Biology and Evolution, Vol. 21, No. 4, Ó Society for Molecular Biology and Evolution 2004; all rights reserved. 632 The single-copy AGT gene has the potential to encode a cleavable N-terminal mitochondrial targeting sequence (MTS) and a C-terminal type 1 peroxisomal targeting sequence (PTS1) by the variable use of two transcription and two translation initiation sites (see figure 3A) (Oda, Funai, and Ichiyama 1990; Oatey, Lumb, and Danpure 1996; Danpure 1997). In many species in which AGT is entirely peroxisomal (e.g., human, rabbit, and guinea pig) translation start site 1 has been lost during evolution (Takada et al. 1990; Purdue, Lumb, and Danpure 1992; Birdsey and Danpure 1998), whereas AGT in the cat is almost entirely mitochondrial because of absence of the second transcription start site (Lumb, Purdue, and Danpure 1994). Species that retain both translation and transcription start sites, such as the rat (Oda, Funai, and Ichiyama 1990) and the marmoset (Purdue, Lumb, and Danpure 1992), retain the potential to target AGT to both organelles. The final intracellular destination of AGT is dependent on the expression of the MTS rather than that of the PTS1, with the former being functionally dominant over the latter (Oatey, Lumb, and Danpure 1996). Thus, a decrease in mitochondrial targeting can lead to an increase in peroxisomal targeting. The only group of mammals in which the evolutionary biology, as opposed to the cell or molecular biology, of AGT targeting has been studied in detail is the Anthropoidea suborder of Primates. Analysis of seven Catarrhini and six Platyrrhini suggested that there was negative selection pressure to conserve the functioning of the MTS in their early evolutionary history but positive selection pressure to diminish or abolish its function in many later branches (Holbrook et al. 2000). Despite the fact that the diets of many extant Primates are not particularly extreme or specialized, most being opportunistic omnivores, the changes to the MTS could be related to a general evolutionary trend of increasing body size and adaptation to diets that were less carnivorous/insectivorous to ones that were more frugivorous and folivorous. Unlike Primates, extreme differences in diet can be found in other mammalian orders such as Carnivora. Molecular Adaptation of AGT Targeting in Carnivora 633 Although most members of the Carnivora are carnivorous, some are omnivorous, and a few, such as the giant panda, are almost entirely herbivorous (Nowak 1991). In the present study, we have investigated whether the evolution of plant-eating in the Carnivora has been accompanied by specific changes to AGT targeting similar to those found in Primates, and whether any changes have resulted from positive evolutionary selection pressure at the molecular level. We have determined the subcellular distribution of AGT in 23 members of Carnivora and analyzed the 59 region of the AGT gene (including both translation start sites and the region encoding the MTS) in seven members with carnivorous, omnivorous, and herbivorous diets. Materials and Methods Animals The following members of Carnivora were investigated or discussed in this study: polecat ferret (Mustela putorius furo), oriental small-clawed otter (Amblonyx cinereus), Chinese ferret badger (Melogale moschata), hog badger (Arctonyx collaris), domestic dog (Canis familiaris), African hunting dog (Lycaon pictus), raccoon dog (Nyctereutes procyonoides), domestic cat (Felis catus), ocelot (Felis pardalis), clouded leopard (Neofelis nebulosa), Persian leopard (Panthera pardus saxicolor), Asiatic lion (Panthera leo persica), Sumatran tiger (Panthera tigris sumatrae), small-toothed palm civet (Arctogalidia trivirgata), slender-tailed meerkat (Suricata suricatta), binturong (Arctictis binturong), blotched genet (Genetta tigrina), mongoose (Herpestes urva), brown bear (Ursus arctos), giant panda (Ailuropoda melanoleuca), sloth bear (Melursus ursinus), raccoon (Procyon lotor), coati (Nasua narica), red panda (Ailurus fulgens), California sea lion (Zalophus californianus), and grey seal (Halichoerus grypus). Frozen blood samples for DNA extraction were obtained from the Zoobank in the Institute of Zoology, London, UK. Liver samples for immunoelectron microscopy were obtained from the pathology tissue archive, Zoological Society of London. Additional samples of giant panda liver were kindly provided by Richard Montali, National Zoological Park, Washington D.C. Samples of tissue were taken at routine necroscopy of animals that had been found dead or from animals shortly after they had been euthanased on humane grounds. Immunoelectron Microscopy Analysis of the intracellular compartmentalization of immunoreactive AGT was determined by immunoelectron microscopy (Cooper et al. 1988; Danpure et al. 1989). In many instances, fresh glutaraldehyde-fixed liver was not available. On these occasions, we made use of the Institute of Zoology’s extensive archived collection of formalinfixed wax-embedded liver samples, which were reprocessed as described previously (Lewin et al. 1995). Liver sections were either immunolabelled singly for AGT using 10 nm gold conjugated goat anti-rabbit IgG, or doubleimmunolabelled for AGT (10 nm gold) followed by the peroxisomal marker catalase (20 nm gold) or the mitochon- drial marker (organelle-specific mitochondrial antigens prepared from homogenized purified rat liver mitochondria) (20 nm gold). As quantitative morphometry was not possible on inevitably scarce and non-ideally fixed specimens, we used the relative number of gold particles per organelle to categorize the distribution of AGT between mitochondria and peroxisomes. If the ratio of the gold labeling density per mitochondrion to that per peroxisome, in specimens singly immunolabeled for AGT, was greater than the arbitrarily chosen value of 10, then the AGT distribution was defined as mitochondrial. If the reverse pertained, then AGT was defined as being peroxisomal. Labeling ratios in between these two extremes led to the distribution being defined as mitochondrial þ peroxisomal. Molecular Biology To obtain species-specific AGT sequence, PCR amplification of genomic DNA from California sea lion, grey seal, brown bear, sloth bear, and giant panda was carried out using degenerate primers P1-CTSMTGYTDGGBCCBGGHCCYTC and P2-CAGBGTGAGBGGGTTCCTGGTCTGGAA that mapped to conserved regions of AGT exons 1 and 2, respectively. Rapid amplification of genomic ends (RAGE) was used to amplify 59-AGT genomic sequence containing both of the potential translation initiation sites, as described previously (Mizobuchi and Frohman 1993; Birdsey and Danpure 1998). Briefly, genomic DNA from selected Carnivora was digested with either restriction enzyme BamHI, EcoRI, or HindIII (Promega). Plasmid pBluescript KSþ (Stratagene) was digested with the same enzymes and then dephosphorylated with calf intestinal alkaline phosphatase (Promega). Digested genomic DNA and plasmid DNA were ligated with T4 DNA ligase (Promega). PCR amplification was carried out using the AGT-specific primer P2 and a vector primer P3-GATGTGTGCAAGGCGATTAAGTTGGT. A second round of PCR amplification was performed using 0.1% of the first amplification products with a nested vector primer P4-CAGGGTTTCCCAGTCACGACGTTT and a nested AGT-specific primer P5-GGCCTACYGGAACATCTCYTTGTG. The final RAGE products were gel purified and cloned into pGEM T-Easy vector (Promega), before sequencing. To analyze the efficiency of mitochondrial import, various AGT N-terminal MTS-GFP fusions were constructed for expression studies in tissue culture cells. PCR was used to amplify the 59 region of the AGT gene flanking the MTS. PCR amplification was carried out on genomic DNA using the primer pairs P6-ACGTAAGCTTACAAGGCCCCCTGGGAAATG and P7-ACGTGGATCCGGCCCGCACCCCACCTACCAC (giant panda) and P6 and P8-ACGTGGATCCGGTCCGCACCCAACCTGCC (sloth bear) and carried out on cDNA clones using P9-ACGTAAGCTTAGGCCAGCTGGGAAATGTTCC and P10CGAGGATCCGGCCTGCCCACCGGCCAC (domestic cat [Lumb, Purdue, and Danpure 1994]) and P11-ACGAAGCTTAGCTCCGGGAATGTTCC and P12-CGAGGATCCGGCTCACCCACCTGCCGC (olive baboon [Holbrook et al. 2000]). PCR products were gel purified and cloned into the HindIII /BamHI sites of pEGFP-N3 (Clontech). 634 Birdsey et al. Cell Culture, Transfection, and Confocal Microscopy SV40-transformed monkey kidney cells (COS1a) were grown on glass coverslips in Dulbecco’s modified Eagle’s medium (Invitrogen), supplemented with 10% fetal calf serum at 378C under 5% CO2. Cells were transfected with the MTS-GFP fusion constructs using Superfect (Qiagen), according to the manufacturer’s instructions. Two days after transfection, the cells were washed in phosphate-buffered saline and stained with the autofluorescent vital dye MitoTracker (Molecular Probes, Cambridge Bioscience). The cells were then fixed in freshly prepared 3% (w/v) paraformaldehyde for 15 min at room temperature, followed by permeabilization with 1% Triton X-100 for 15 min at room temperature. The cover slips were mounted onto glass slides in Mowiol (Harlow Chemical Co. Ltd) containing diazabicyclo[2,2,2]octane (DABCO, Sigma). The autofluorescent staining pattern of GFP and MitoTracker was visualized in a BioRad MRC1024 confocal laser-scanning microscope and the images were digitally recorded. Data Analysis Nucleotide sequences were aligned using ClustalW version 1.7 (Thompson, Higgins, and Gibson 1994). Reconstruction of ancestral sequences was carried out by codon-based maximum-likelihood analysis using marginal reconstruction of ancestral sequences (Yang, Kumar, and Nei 1995). Estimation of the dN/dS(x) among lineages was calculated by the maximum-likelihood method (Yang 1998) using the computer software package PAML version 3.13a (Yang 1997). In all analyses, the codon frequencies were calculated from the average nucleotide frequencies (F1X4), and the transition/transversion rate ratio (kappa) was fixed at 1. Likelihood ratio tests were used to compare models, with the v2 distribution used as an approximation. Computer prediction of the targeting efficiency of AGT MTSs was carried out using PSORT (Nakai and Horton 1999). Results We determined the subcellular distribution of AGT using immunoelectron microscopy on liver sections from 20 species from the order Carnivora (figs. 1 and 2A). These results, together with those already known for the domestic cat, domestic dog, and polecat ferret (Danpure et al. 1994), were compared with their natural diets (fig. 2A and B). Of the 13 species classified as mainly or entirely carnivorous, all but one (mongoose) had AGT mainly or exclusively in the mitochondria (fig. 1A and B). At the other extreme, all three mainly herbivorous species (coati, red panda, and giant panda) had AGT in both mitochondria and peroxisomes (figs. 1C, 1D, and 2A). In all but two of the eight species categorized as omnivorous, AGT was also both mitochondrial and peroxisomal. Although the distribution of AGT was invariant in some families, such as Canidae and Felidae (mitochondrial) and Procyonidae (mitochondrial and peroxisomal), in others, such as Mustelidae (mitochondrial or mitochondrial and peroxisomal), it was variable. Unlike many members of other orders (Danpure et al. 1994), in no cases were any Carnivora found to have only peroxisomal AGT. Nevertheless, there was a clearly identifiable trend in which carnivorous Carnivora were more likely to have mitochondrial AGT, whereas omnivorous and herbivorous Carnivora were more likely to have both mitochondrial and peroxisomal AGT (fig. 2B). To determine the molecular basis for the changes in AGT distribution in Carnivora, we analyzed the AGT gene from seven species: domestic cat (Felis catus), blotched genet (Genetta tigrina), California sea lion (Zalophus californianus), grey seal (Halichoerus grypus), brown bear (Ursus arctos), sloth bear (Melursus ursinus), and giant panda (Ailuropoda melanoleuca). Using genomic DNA as template, the 59 region of AGT was amplified by PCR and RAGE (Mizobuchi and Frohman 1993) (with the exception of the domestic cat, the cDNA of which had previously been cloned [Lumb, Purdue, and Danpure 1994]). The amplified region included all the sequence from exon 1 and most or all of the 59 UTR (see fig. 3A). Analysis of the sequences revealed the presence of both ancestral translation start sites (1 and 2 in figure 3A) in all seven species, as predicted from the observation that at least some AGT is mitochondrial in all cases. An alignment (fig. 3B) of the 55–amino acid sequence encoded by exon 1 downstream of, and including, the second translation start site revealed a high degree of conservation with an 84% sequence identity across all seven species. In contrast, a similar analysis of the 22–amino acid sequence (19 amino acids in the sloth bear) with potential to encode the MTS (i.e., the region upstream of the second translation start site) showed only 50% amino acid identity. Such low conservation is unsurprising as MTSs are defined more by their three-dimensional structure (positively-charged amphiphilic a-helices [von Heijne 1986]) than by any particular consensus sequence. To determine the number of changes that have occurred to the MTS during the evolution of Carnivora and their temporal relationships, we used a codon-based maximum-likelihood analysis (Yang, Kumar, and Nei 1995) to reconstruct ancestral sequences (fig. 4A). The accuracy of the reconstruction was maximized by the inclusion of the N-terminal sequences of 24 additional mammals, including those from the Anthropoidea previously analyzed (Holbrook et al. 2000) and a variety of others, both published and unpublished (see legend to figure 4A for details). In the Carnivora, by far the most amino acid substitutions occurred in the most recent branch leading to the giant panda. A third (7/22) of the amino acids comprising the MTS had changed in this one branch. Many of the changes in the giant panda MTS are predicted to decrease the effectiveness of the MTS by decreasing its helix-forming potential and decreasing its positive charge. Other branches have far fewer or no amino acid changes and, with the exception of the sloth bear, are not predicted to have any influence on the effectiveness of the MTS. The sloth bear lineage is unusual in that there has been a loss of three adjacent amino acids, the consequences of which are less predictable. Analysis of the MTS region using the PSORT computer program (Nakai and Horton 1999) predicts that the MTS of the Molecular Adaptation of AGT Targeting in Carnivora 635 FIG. 1.—Dietary dependence of AGT distribution in the livers of Carnivora. Subcellular distribution of AGT was determined by immunoelectron microscopy in liver sections obtained from necroscopy specimens from (A) oriental small-clawed otter (carnivore), (B) blotched genet (carnivore), (C) red panda (herbivore-omnivore), and (D) giant panda (herbivore). In A, B, C, and D1, the specimens were singly labeled for AGT (10 nm gold). In D2, the specimen was doubly immunolabeled for AGT (10 nm gold) and the peroxisomal marker catalase (20 nm). In D3, the specimen was doubly immunolabeled for AGT (10 nm gold) and organelle-specific mitochondrial antigens (20 nm). M ¼ mitochondrion, P ¼ peroxisome. The number in brackets after M or P indicates the number of 10 nm gold particles located in the cross-sectional profile of that organelle (for comparative purposes only). For the giant panda (D) the areas indicated by the rectangles in the upper images are magnified to give the lower images. Size bars ¼ 200 nm. Such immunolabeling shows that AGT is mitochondrial in the carnivores (A and B) but both peroxisomal and mitochondrial in the herbivores (C and D). giant panda and the sloth bear should be very weak, whereas that of the blotched genet, domestic cat, brown bear, grey seal, and California sea lion should be strong (fig. 6A). Interestingly, the PSORT prediction of the reconstructed ancestral mammalian MTS (fig. 4A) suggested that it would have a lower mitochondrial targeting efficiency than the MTS of the ancestral Carnivora (1.09 compared to 1.49, respectively). Ancestral reconstruction of exon 1 sequence downstream of the MTS in the Carnivora (fig. 4B) showed a very different picture to that found in the MTS region. Although several amino acid changes were identified in a number of branches, the most recent branch leading to the giant panda contained no changes at all, even though this region contained 55 amino acids (compared to 22 amino acids in the MTS region). Amino acid changes in the MTS region could be neutral or an adaptive response to selection pressure to change AGT distribution. Using a maximum-likelihood method (Yang 1998), we have determined the relative number of nonsynonymous differences per nonsynonymous site (dN) compared with the number of synonymous differences per synonymous site (dS) in the region with potential to encode a MTS and downstream of this region. The dN/dS ratio (x) provides a measure of the selection pressure acting on genes or parts of genes. Thus, dN/dS , 1 suggests that there has been negative selection pressure (i.e., pressure for conservation), dN/dS . 1 suggests that 636 Birdsey et al. FIG. 2.—AGT distribution in Carnivora is related to diet. (A) Phylogenetic tree (Bininda-Emonds, Gittleman, and Purvis 1999) of 26 members of Carnivora showing natural diet (Carniv ¼ carnivorous, Omniv ¼ omnivorous, Herbiv ¼ herbivorous, Carniv-Omniv ¼ mainly carnivorous, but some plant material eaten, Herbiv-Omniv ¼ mainly herbivorous, but some animal material eaten) and the subcellular distribution of AGT in liver cells (M ¼ mitochondrial, MþP ¼ mitochondrial plus peroxisomal, ? ¼ unknown). (B) Relationship between natural diet and intracellular compartmentalization of AGT in Carnivora. All positions within any one box are equivalent. Numbering is the same as for figure 2A. As no members of Carnivora have yet been categorized as having only peroxisomal AGT, a few species from other orders have been included for comparison. Herbiv ¼ herbivorous, Omniv ¼ omnivorous, Carniv ¼ carnivorous. there has been positive selection pressure (i.e., pressure for change), and dN/dS ¼ 1 suggests that the region is neutral (i.e., no selection pressure). When comparing the MTS region in the seven Carnivora (fig. 5A), we find that out of the branches where dS 6¼ 0, dN/dS . 1 in only one (i.e., the most recent branch leading to the giant panda). In all other branches where dS 6¼ 0, dN/dS , 1. In the one other Carnivora branch where dN/dS . 1 (i.e., branch 10), there has been one nonsynonomous substitution and no synonomous substitutions, giving dN/dS ¼ ‘. The significance Molecular Adaptation of AGT Targeting in Carnivora 637 FIG. 3.—(A) Diagrammatic structure of the archetypal mammalian AGT gene. Ancestral translation start sites 1 and 2 and transcription start sites A and B, are indicated. The region encoding the MTS (between translation start sites 1 and 2) and the PTS1 are shown. (B) Deduced amino acid sequences encoded by the 59 region of the AGT gene in various Carnivora. In the aligned sequences, asterisks denote residues identical between all species. The amino acids encoded by the two translation sites are indicated in bold. of this observation is unclear and may be caused by a threshold effect associated with the comparison of short sequences (see Holbrook et al. [2000]). To determine whether the dN/dS ratio in the lineage leading to the giant panda was significantly different (i.e., greater) than in other Carnivora lineages, the Carnivora data set was fitted to various models of sequence evolution. The models were compared by the likelihood ratio test using the v2 approximation (Yang 1998). The ‘‘one-ratio’’ model (which assumes the same ratio for all branches in the phylogeny) was compared with the ‘‘two-ratio’’ model (which assumes that the branch of interest has a dN/dS ratio that is different from the background ratio). The loglikelihood value from the two-ratio model is l1 ¼191.89. The log-likelihood value obtained under the one-ratio model is l0 ¼194.11. Twice the log-likelihood difference, 2l ¼ (l1 l0), can be compared with a v2 distribution to test whether the two-ratio model fits the data significantly ! FIG. 4.—Ancestral reconstruction of the amino acid substitutions in the N-terminus of mammalian AGT. Reconstructions were carried out using a codon-based maximum-likelihood analysis (Yang, Kumar, and Nei 1995). The phylogenetic tree is based on that of Springer et al. (2003) and includes all mammalian species (31 in total) in which the N-terminal sequence of AGT is known. The branches are arbitrarily labeled 1 to 59. (A) MTS region; (B) exon 1 downstream of the MTS region. The ancestral node is indicated by the closed circle and the calculated ancestral sequence is indicated to the left of the tree. The amino acid substitutions are shown along each branch. The asterisk (*) in branch 34 (slow loris) in (A) indicates an insertion of R between residues 3 and 4. In addition to the seven members of Carnivora, the following species are also shown on the tree: brown big-eared bat (Plecotus auritus), domestic cow (Bos taurus), pigmy hippopotamus (Hexaprotodon liberiensis), harbor porpoise (Phocena phocoena), killer whale (Orcinus orca), Przewalski’s horse (Equus caballus przewalskii), European rabbit (Oryctolagus cuniculus), guinea pig (Cavia porcellus), Norway rat (Rattus norvegicus), house mouse (Mus musculus), slow loris (Nycticebus coucang), white-handed gibbon (Hylobates lar), common gorilla (Gorilla gorilla), human (Homo sapiens), chimpanzee (Pan troglodytes), Diana monkey (Cercopithecus diana), olive baboon (Papio anubis), celebes macaque (Macaca nigra), squirrel monkey (Saimiri sciureus), Goeldi’s monkey (Callimico goeldii), golden lion tamarin (Leontopithecus rosalia), common marmoset (Callithrix jacchus), white-faced saki monkey (Pithecia pithecia), and black spider monkey (Ateles paniscus). The N-terminal AGT sequences used in the PAML analysis were obtained from the following sources: cow (Takasuga et al. 2001), rabbit (Purdue, Lumb, and Danpure 1992), cat (Lumb, Purdue, and Danpure 1994), guinea pig (Birdsey and Danpure 1998), rat (Oda et al. 1987), mouse (Li, Salido, and Shapiro 1999), and Primates (Holbrook et al. 2000). The remaining sequences were obtained from genomic DNA using the RAGE technique (see Materials and Methods). Accession numbers for the unpublished AGT sequences are listed as Supplementary Material. The Carnivora analyzed as part of the present study and the Anthropoidea analyzed previously by Holbrook et al. (2000) are boxed in gray. In (A), branches containing loss of translation start site 1 are dashed, to indicate that at some point along the branch, the MTS would have been excluded from the open reading frame. 638 Birdsey et al. Molecular Adaptation of AGT Targeting in Carnivora 639 FIG. 4. (Continued). 640 Birdsey et al. better than the one-ratio model. The test statistic 2l ¼ 4.44 has a P value of 0.035, which indicates that the dN/dS ratio for the MTS region in the branch leading to the giant panda is significantly greater than that found in other Carnivora branches of the tree. To test whether the dN/dS ratio for the giant panda lineage (x1) was significantly greater than 1, a likelihood ratio test was carried out on the Carnivora data set using the two-ratio model with and without x1 fixed at 1. The log-likelihood value obtained when x1 was estimated by maximum likelihood is l1 ¼ 191.89, compared with a value of l0 ¼ 192.66 when x1 ¼ 1. The test statistic 2l ¼ 1.54 with P ¼ 0.215 indicates that the difference between the two models was not significant. Notwithstanding this latter result, which in itself is not incompatible with the existence of positive selection pressure (Yang 1998), all the data taken together are compatible with the suggestion that, within Carnivora, there has been positive selection pressure to alter the MTS in the giant panda lineage. In constrast, evolution of this region in most other Carnivora lineages appears to have been subjected to negative selection pressure (i.e, pressure to maintain its function as a MTS). Although not part of the present study, it is interesting to note that analysis of the dN/dS ratio in a number of other mammalian lineages also show some branches with x greater than 1 (fig. 5A). This would be compatible with episodic adaptive evolution of the MTS of AGT across Mammalia, similar to that identified in primate lysozymes (Messier and Stewart 1997). When the rest of exon 1 (i.e., downstream of the MTS) is compared in the different Carnivora species, a rather different picture emerges. In this region dN/dS , 1 in all branches in which dS 6¼ 0 (fig. 5B). Of particular note is the fact that the giant panda branch contains no nonsynonymous mutations, and only one synonymous mutation, in this region of 165 nucleotides (55 amino acids), whereas there were seven nonsynonymous changes in the MTS region of 66 nucleotides (22 amino acids) (see above). Although the effectiveness of the MTSs of the giant panda and the sloth bear were predicted to be much less than those of the more-carnivorous species, such as the domestic cat and blotched genet, using the PSORT computer program (fig. 6A), a direct measurement of their strength was made using a mitochondrial-reporter protein import assay in tissue culture cells. Constructs containing the MTS from different species (domestic cat, sloth bear, and giant panda) were attached to the N-terminus of green fluorescent protein (GFP) and transiently expressed in COS1a tissue culture cells. The targeting of the fusion constructs was determined by confocal fluorescence microscopy (fig. 6B–F). GFP alone and GFP fused to the ‘‘MTS’’ of baboon (Papio anubis) AGT (Holbrook et al. 2000), which has previously been shown not to target to mitochondria (Danpure et al. 1990) and which has no features of an MTS as determined by the PSORT program (fig. 6A), were used as negative controls. In the absence of any mitochondrial targeting, GFP and baboon MTS-GFP were diffusely distributed across the cytosol and nucleus, as is frequently found in nontargeted GFP constructs (Lumb, Drake, and Danpure 1999) (see also http://www. clontech.com/techinfo/faqs/LivingColors/general.shtml). In contrast, the MTS of domestic cat AGT was very efficient at targeting GFP, all of which appeared to be mitochondrial, as shown by the yellow colocalization of GFP with the mitochondrial marker MitoTracker and the absence of any GFP in the cytosol or nucleus (fig. 6D). However, the ability of the MTSs of sloth bear and giant panda to target GFP fusions to mitochondria was much less (fig. 6E and F). In both cases, although some mitochondrial localization of the fusion proteins was observed, most failed to target and was cytosolic and nuclear. These targeting studies are compatible with the predictions of the PSORT analysis, that the mutations that occur in the MTS of sloth bear and giant panda decrease its effectiveness. At least in the case of the giant panda, this finding is entirely compatible with the previous evidence ! FIG. 5.—Nonsynonymous/synonymous substitution ratios (dN/dS) among lineages in the 59-region of the AGT gene in mammals. The phylogenetic trees were derived from that of Springer et al. (2003) using the PAML program (see Material and Methods) and drawn using the TREEVIEW program (Page 1996). The branch lengths and the dN/dS ratios were calculated using the free-ratios model (Yang 1998). The apparent loss of some branches results from the fact that in some cases dN ¼ dS ¼ 0, so that the branch length is zero. (A) MTS region; (B) exon 1 downstream of the MTS. The animals studied and their sources are described in the legend to figure 4. The Carnivora analyzed as part of the present study and the Anthropoidea analyzed previously by Holbrook et al. (2000) are boxed in gray. Branches in which dN/dS . 1 are shown by the use of thick lines. In (A), branches containing loss of translation start site 1 are dashed, to indicate that at some point along the branch, the MTS would have been excluded from the open reading frame. In branches where dN/dS . 1, the ratio is shown above the branch. An asterisk (*) indicates that dS ¼ 0, so that dN/dS ¼ ‘. The calculated dN/dS ratios (dN, dS) along the various branches are as follows: MTS (A) 1 ¼ 0.021 (dN), 0.106 (dS); 2 ¼ 0, 0; 3 ¼ 0.02, 0.05; 4 ¼ 0, 0; 5 ¼ 0, 0; 6 ¼ 0.165, 0.051; 7 ¼ 0, 0; 8 ¼ 0, 0.056; 9 ¼ 0, 0; 10 ¼ 0.021, 0; 11 ¼ 0.043, 0.106; 12 ¼ 0.042, 0.109; 13 ¼ 0.034, 0.055; 14 ¼ 0.069, 0.327; 15 ¼ 0.006, 0.048; 16 ¼ 0.077, 0.048; 17 ¼ 0.021, 0; 18 ¼ 0.113, 0.195; 19 ¼ 0, 0; 20 ¼ 0.051, 0.379; 21 ¼ 0, 0; 22 ¼ 0, 0; 23 ¼ 0.02, 0; 24 ¼ 0.122, 0.308; 25 ¼ 0.045, 0.05; 26 ¼ 0, 0; 27 ¼ 0.247, 0.079; 28 ¼ 0, 0; 29 ¼ 0.181, 0.299; 30 ¼ 0.123, 0.39; 31 ¼ 0, 0; 32 ¼ 0.046, 0.204; 33 ¼ 0, 0.05; 34 ¼ 0.209, 0.158; 35 ¼ 0.022, 0.099; 36 ¼ 0.024, 0.051; 37 ¼ 0, 0; 38 ¼ 0.049, 0.045; 39 ¼ 0.05, 0.045; 40 ¼ 0.025, 0; 41 ¼ 0, 0; 42 ¼ 0, 0; 43 ¼ 0.026, 0; 44 ¼ 0.075, 0; 45 ¼ 0.101, 0; 46 ¼ 0, 0; 47 ¼ 0, 0; 48 ¼ 0, 0; 49 ¼ 0, 0.218; 50 ¼ 0, 0; 51 ¼ 0.076, 0; 52 ¼ 0, 0; 53 ¼ 0, 0.049; 54 ¼ 0, 0; 55 ¼ 0.05, 0; 56 ¼ 0, 0; 57 ¼ 0, 0; 58 ¼ 0.076, 0; 59 ¼ 0.051, 0. Exon 1 (B) 1 ¼ 0 (dN), 0.039 (dS); 2 ¼ 0, 0.032; 3 ¼ 0.009, 0.113; 4 ¼ 0, 0.115, 5 ¼ 0.009, 0.049; 6 ¼ 0, 0.022; 7 ¼ 0, 0; 8 ¼ 0.009, 0; 9 ¼ 0.017, 0; 10 ¼ 0, 0.127; 11 ¼ 0.064, 0.129; 12 ¼ 0.009, 0.044; 13 ¼ 0, 0.063; 14 ¼ 0.042, 0.131; 15 ¼ 0, 0; 16 ¼ 0.056, 0.106; 17 ¼ 0.017, 0; 18 ¼ 0.066, 0.092; 19 ¼ 0.013, 0.156; 20 ¼ 0.029, 0.134; 21 ¼ 0, 0; 22 ¼ 0, 0.071; 23 ¼ 0, 0.08; 24 ¼ 0.025, 0.176; 25 ¼ 0, 0; 26 ¼ 0, 0.014; 27 ¼ 0.067, 0.4; 28 ¼ 0, 0; 29 ¼ 0.042, 0.284; 30 ¼ 0.074, 1.23; 31 ¼ 0.039, 0.208; 32 ¼ 0, 0; 33 ¼ 0, 0.055; 34 ¼ 0.059, 0.161; 35 ¼ 0.05, 0.109; 36 ¼ 0.022, 0.079; 37 ¼ 0.013, 0.022; 38 ¼ 0.026, 0.117; 39 ¼ 0.035, 0.037; 40 ¼ 0, 0; 41 ¼ 0, 0; 42 ¼ 0, 0; 43 ¼ 0, 0; 44 ¼ 0.043, 0; 45 ¼ 0.099, 0.087; 46 ¼ 0, 0.116; 47 ¼ 0.018, 0.09; 48 ¼ 0.009, 0.038; 49 ¼ 0.016, 0.052; 50 ¼ 0, 0; 51 ¼ 0.051, 0.091; 52 ¼ 0.016, 0.057; 53 ¼ 0, 0.047; 54 ¼ 0, 0; 55 ¼ 0, 0; 56 ¼ 0.008, 0.126; 57 ¼ 0, 0.041; 58 ¼ 0.033, 0.06; 59 ¼ 0.096, 0.099. For the purposes of this analysis, the R inserted between residues 3 and 4 in the slow loris MTS was removed and the alignment gap created by the deletion of residues F2, R3, and A4 in the sloth bear MTS was not taken into account. The results of the analysis of Anthropoidea are slightly, but not significantly, different from those obtained by Holbrook et al. (2000). This is because of the inclusion in the current, but not the previous, analysis of translation start site 1 and the need to change some of the program parameters to optimize the analysis over many more species. Molecular Adaptation of AGT Targeting in Carnivora 641 642 Birdsey et al. FIG. 5. (Continued). Molecular Adaptation of AGT Targeting in Carnivora 643 presented in this paper that positive selection pressure for change has led to decreased mitochondrial targeting of AGT and, hence, its increased peroxisomal targeting. Although the subcellular distribution of AGT in the sloth bear is currently unknown, the deletion of three amino acids from the MTS and the inefficient mitochondrial import of the sloth bear MTS-GFP fusion protein are suggestive of a similar AGT distribution to that seen in the giant panda (i.e., mitochondrial and peroxisomal). The extent to which this represents an adaptive response to the unusual seasonally herbivorous diet of the sloth bear (Nowak 1991; Joshi, Garshelis, and Smith 1997) remains to be determined. Discussion In the present study, we have extended our analyses of the adaptive evolution of AGT targeting in mammals under the influence of dietary selection pressure, by determining the subcellular distribution of AGT in the livers of Carnivora, the members of which have extremely divergent diets, ranging from absolute carnivory in lions and tigers to absolute herbivory in the giant panda. In addition, we have investigated the evolutionary changes to the structure and function of the region of AGT known to be responsible for its targeting to mitochondria (i.e., its Nterminus). The results show that the distribution of AGT in Carnivora is related to diet, as it is in Mammalia as a whole, there being an adaptive shift in its distribution from mitochondria to peroxisomes as diets change from being more carnivorous to more herbivorous. Thus, in the most carnivorous species, AGT is more likely to be entirely or mainly mitochondrial, whereas in most omnivorous or herbivorous species, AGT is more likely to be spread more evenly between both mitochondria and peroxisomes. Although, unlike many other mammals, no members of Carnivora had exclusively peroxisomal AGT, the greatest proportion of peroxisomal AGT was found in the most herbivorous member of the order, the giant panda. The results also show that the shift in AGT targeting away from the mitochondria and toward the peroxisomes in the giant panda is related to the accumulation of nonsynonymous mutations in the MTS that decrease its efficiency compared with that in carnivorous Carnivora. In addition, evidence is presented that the MTS in the most recent lineage leading to the giant panda has been subjected to positive selection pressure to change (i.e., to decrease its efficiency), thereby allowing more AGT to be targeted to the peroxisomes. This could have been a defining moment in the evolution of the most herbivorous member of the order, enabling the giant panda to detoxify glyoxylate derived from plant-based oxalate precursors, such as glycolate, with greater efficiency at the expense of a lower efficiency for the detoxification of glyoxylate derived from meat-based precursors, such as hydroxyproline. AGT distribution has changed on numerous occasions during the evolution of mammals (see Introduction), but only in the case of Primates has it been suggested to be the result of positive selection pressure (Holbrook et al. 2000). Where a temporal trend can be discerned, the change in AGT distribution in mammals is manifested by an increase in peroxisomal AGT at the expense of mitochondrial AGT. No clear-cut examples of the reverse have yet been found. More particularly, once mitochondrial AGT targeting has been lost, it is unlikely to be reacquired (one notable example where this is not the case is found in humans homozygous for a normally occurring Pro11 Leu polymorphism, which creates a weak MTS sufficient to target approximately 5% of their AGT to mitochondria (Danpure 1997)). This fits in with the general trend away from carnivory and toward herbivory that characterises mammalian evolution in many lineages. Although there are similarities between the evolution of AGT targeting in Carnivora and Primates (Holbrook et al. 2000), there are significant differences too. These differences may reflect differences in the nature of the dietary selection pressure in the two orders. For example, although there has been a general shift away from more carnivorous/insectivorous diets toward more frugivorous/ folivorous diets in Primates, within Carnivora some lineages (e.g., giant panda) have moved in the direction of herbivory, whereas others (e.g., lions, tigers etc) have probably become more carnivorous relative to the ancestral Carnivora. In this respect, it is interesting to note that the PSORT mitochondrial targeting index of the reconstructed ancestral AGT MTSs varies significantly. For example, the PSORT indices for the MTS in the ancestral mammal (sequence at the ancestral node in figure 4A), the ancestral Carnivora (branch 13 in figure 4A), and the extant cat (branch 11) are 1.09, 1.49, and 1.79, respectively. This is compatible with increasing carnivory and increasing mitochondrial AGT. The index for the brown bear at 1.49 might suggest a diet and AGT distribution similar to that of the ancestral Carnivora. On the other hand, the index for the giant panda at 1.24 is compatible with a diet much more herbivorous than that of the ancestral Carnivora and much more AGT in the peroxisomes. As far as AGT targeting is concerned, the main difference between Carnivora and Primates is that AGT is either mitochondrial or mitochondrial þ peroxisomal in the former, whereas it is either mitochondrial þ peroxisomal or peroxisomal in the latter. AGT has yet to be found exclusively in the peroxisomes in Carnivora or exclusively in the mitochondria in Primates. Whether this is related to the rarity of obligate herbivores in Carnivora and obligate carnivores in Primates is unclear. As far as the molecular basis of changes to AGT targeting are concerned, the main difference between Carnivora and Primates is that whereas mitochondrial AGT targeting is diminished by the accumulation of mutations in the MTS that decrease its efficiency in the former, mitochondrial targeting in Primates has been frequently abolished by loss of translation start site 1 (see Introduction) and permanent exclusion of the MTS from the open reading frame. There is evidence that in one Primate lineage ancestral to the olive baboon and celebes macaque (branch 44 in figure 4A) mitochondrial AGT targeting has been lost also by the accumulation of mutations in the MTS region, as occurs in Carnivora (but see below). Although the differential targeting of AGT between mitochondria and peroxisomes is an unparalleled system 644 Birdsey et al. FIG. 6.—Dietary selection pressure leads to the evolution of an inefficient MTS. (A) PSORT computer prediction scores indicate the relative strength of the MTS from each animal, which are color-coded according to diet (carnivorous species are color-coded red, herbivorous species are green, and omnivorous species are blue). The nonfunctional ‘‘MTS’’ from olive baboon AGT is included for comparison. (B–F) COS cells were transiently transfected with various GFP constructs—GFP alone (B), baboon MTS-GFP (C), domestic cat MTS-GFP fusion (D), sloth bear MTS-GFP fusion (E), and giant panda MTS-GFP fusion (F). GFP and baboon MTS-GFP were used as negative controls. GFP autofluorescence ¼ green, MitoTracker (mitochondrial marker) ¼ red, colocalization ¼ yellow. All panels are at the same magnification. Bar ¼ 10 lm. with which to study the ability of mammals, both extant and ancestral, to adapt to changes in dietary selection pressure, the system is not without its technical difficulties that, to some extent, compromise its robustness. First, the sequence responsible for mitochondrial targeting and subject to dietary selection pressure is short (i.e., only 22 amino acids in most mammals). Second, MTSs tend to be conserved at the structural level (positively charged amphiphilic ahelices) rather than sequence level. This not only makes long distance comparisons (e.g., between different phyla) difficult because the sequences cannot be aligned, but also means that the MTS sequence is less well conserved than most of the rest of the protein, irrespective of any dietary differences. Third, to act as a MTS, the region between translation start sites 1 and 2 (see Introduction) must be contained within the open reading frame. This means that not only must translation start site 1 exist, but also transcription start site A must exist. The former can be determined by sequencing genomic DNA, but the latter can only be determined by transcript mapping, a procedure that requires fresh liver tissue. Although this is no problem with laboratory species, such as the rat, mouse, cat, or even human, it is difficult to obtain such material from wild animals, even those in zoos. In the case of at least six Primates (see figure 4A), it is clear that the MTS is excluded from the open reading frame because the loss of translation start site 1 simply by sequencing genomic DNA (Holbrook et al. 2000). Equally, it is likely that in all the Carnivora studied, the MTS is within the open reading frame, because not only is translation start site 1 present but also AGT is at least partly targeted to mitochondria in all cases. However, in species where translation start site 1 is still present, yet AGT is exclusively targeted to peroxisomes (e.g., as occurs in the baboon), then difficulties can arise. It so happens that in the case of the baboon its MTS does not work (see figure 6) whether it is in the open reading frame or not. Because the present study is based on genomic DNA, rather than cDNA, the big unknown is what is the relative usage of transcription start sites A and B in the AGT gene (see figure 3A) in the members of Carnivora and to what extent this might influence the intracellular distribution of AGT. The only information currently available comes from studies on the domestic cat (Lumb, Purdue, and Danpure 1994), in which all transcripts initiate from site A, upstream of both translation start sites 1 and 2. If this occurred throughout Carnivora, then peroxisomal targeting of AGT could be dependent either on leaky ribosome scanning through translation start site 1, with subsequent translation from start site 2 (see figure 3A), or by a functional weakening of the MTS counteracting its hierarchical dominance over the PTS1 (see Introduction). The molecular determinants of leaky ribosome scanning are unclear, but they might be related to the degree of match of each potential translation start site to the so-called Kozak consensus sequence (Kozak 1986, 1989). In this respect it is interesting to note that the Kozak sequence found at both Molecular Adaptation of AGT Targeting in Carnivora 645 sites are very similar between the domestic cat and the giant panda, even though the latter targets a greater proportion of its AGT to peroxisomes than does the former. Evidence presented above suggests that a functional weakening of the MTS is the more likely explanation. Molecular adaptation of many other genes to dietary changes must have occurred during the evolution of mammals. However, surprisingly few examples have been demonstrated to result in functional changes to the gene product or to be the result of positive selection pressure. A notable and prototypical exception is that of lysozyme that has evolved independently in langurs (Primates) and Artiodactyla to survive the rigours of a nutritionally poor folivorous diet and the necessity of foregut fermentation (Stewart, Schilling, and Wilson 1987; Messier and Stewart 1997). Unlike lysozyme, however, the relationship between AGT distribution and diet is spread across many orders of mammals. If the distribution of AGT is an important determinant of an animal’s diet and if, as seems likely, mitochondrial AGT targeting is more easily lost than acquired, then it is not surprising that secondary carnivory is a rare event in mammalian evolution. Supplementary Material The novel sequences discussed in this paper have been submitted to the Nucleotide Sequence Database under accession numbers AJ539309 (H. grypus.), AJ539310 (Z. californianus), AJ539311 (A. melanoleuca), AJ539312 (G. tigrina), AJ539313 (M. ursinus), AJ539314 (U. arctos), AJ567378 (P. auritus), AJ567379 (O. orca), AJ567380 (E. przewalskii), AJ567381 (H. liberiensis), AJ567382 (P. phocoena), and AJ567383 (N. coucang). Acknowledgments We thank Gemma Athorn for technical assistance. Access to the Zoobank frozen tissue and blood archive was arranged by Dada Gottelli, Institute of Zoology, London. We thank Richard Montali, National Zoological Park, Washington D.C. for providing samples of giant panda liver. We are grateful to Ziheng Yang for help in using the PAML program and to Andy Purvis for helpful discussions. This work was supported by the Leverhulme Trust. Literature Cited Bininda-Emonds, O. R. P., J. L. Gittleman, and A. Purvis. 1999. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia). Biol. Rev. 74:143–175. Birdsey, G. M. and C. J. Danpure. 1998. Evolution of alanine:glyoxylate aminotransferase intracellular targeting. Structural and functional analysis of the guinea pig gene. Biochem. J. 331:49–60. Cooper, P. J., C. J. Danpure, P. J. Wise, and K. M. Guttridge. 1988. Immunocytochemical localization of human hepatic alanine: glyoxylate aminotransferase in control subjects and patients with primary hyperoxaluria type 1. J. Histochem. Cytochem. 36:1285–1294. Danpure, C. J. 1997. Variable peroxisomal and mitochondrial targeting of alanine: glyoxylate aminotransferase in mammalian evolution and disease. Bioessays 19:317–326. ———. 2001. Primary hyperoxaluria. Pp. 3323–3367 in C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler, and B. Vogelstein, eds. The molecular and metabolic bases of inherited disease. McGraw-Hill, New York. Danpure, C. J., P. J. Cooper, P. J. Wise, and P. R. Jennings. 1989. An enzyme trafficking defect in two patients with primary hyperoxaluria type 1: peroxisomal alanine/glyoxylate aminotransferase rerouted to mitochondria. J. Cell Biol. 108:1345– 1352. Danpure, C. J., P. Fryer, P. R. Jennings, J. Allsop, S. Griffiths, and A. Cunningham. 1994. Evolution of alanine:glyoxylate aminotransferase 1 peroxisomal and mitochondrial targeting: a survey of its subcellular distribution in the livers of various representatives of the classes Mammalia, Aves and Amphibia. Eur. J. Cell Biol. 64:295–313. Danpure, C. J., K. M. Guttridge, P. Fryer, P. R. Jennings, J. Allsop, and P. E. Purdue. 1990. Subcellular distribution of hepatic alanine:glyoxylate aminotransferase in various mammalian species. J. Cell Sci. 97:669–678. Danpure, C. J. and L. H. Smith. 1996. The primary hyperoxalurias. Pp. 859–881 in F. L. Coe, M. J. Favus, C. Y. Pak, J. H. Parks, and G. M. Preminger, eds. Kidney stones: medical and surgical management. Lippincott-Raven, Philadelphia. Holbrook, J. D., G. M. Birdsey, Z. Yang, M. W. Bruford, and C. J. Danpure. 2000. Molecular adaptation of alanine:glyoxylate aminotransferase targeting in primates. Mol. Biol. Evol. 17:387–400. Joshi, A. R., D. L. Garshelis, and J. L. D. Smith. 1997. Seasonal and habitat-related diets of sloth bears in Nepal. J. Mammalogy 78:584–597. Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292. ———. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229–241. Lewin, J., A. P. Dhillon, R. Sim, G. Mazure, R. E. Pounder, and A. J. Wakefield. 1995. Persistent measles virus infection of the intestine: confirmation by immunogold electron microscopy. Gut 36:564–569. Li, X. M., E. C. Salido, and L. J. Shapiro. 1999. The mouse alanine:glyoxylate aminotransferase gene (Agxt1): cloning, expression, and mapping to chromosome 1. Somat. Cell Mol. Genet. 25:67–77. Lumb, M. J., A. F. Drake, and C. J. Danpure. 1999. Effect of Nterminal alpha helix formation on the dimerization and intracellular targeting of alanine:glyoxylate aminotransferase. J. Biol. Chem. 274:20587–20596. Lumb, M. J., P. E. Purdue, and C. J. Danpure. 1994. Molecular evolution of alanine/glyoxylate aminotransferase 1 intracellular targeting: analysis of the feline gene. Eur. J. Biochem. 221:53–62. Messier, W., and C. B. Stewart. 1997. Episodic adaptive evolution of primate lysozymes. Nature 385:151–154. Mizobuchi, M., and L. A. Frohman. 1993. Rapid amplification of genomic DNA ends. Biotechniques 15:214–216. Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends. Biochem. Sci. 24:34–36. Noguchi, T. 1987. Amino acid metabolism in animal peroxisomes. Pp. 234–243 in H. D. Fahimi and H. Sies, eds. Peroxisomes in biology and medicine. Springer-Verlag, Berlin. Nowak, R. M. 1991. Walker’s mammals of the world. John Hopkins University Press, Baltimore. Oatey, P. B., M. J. Lumb, and C. J. Danpure. 1996. Molecular basis of the variable mitochondrial and peroxisomal 646 Birdsey et al. localization of alanine:glyoxylate aminotransferase. Eur. J. Biochem. 241:374–385. Oda, T., T. Funai, and A. Ichiyama. 1990. Generation from a single gene of two mRNAs that encode the mitochondrial and peroxisomal serine:pyruvate aminotransferase of rat liver. J. Biol. Chem. 265:7513–7519. Oda, T., H. Miyajima, Y. Suzuki, and A. Ichiyama. 1987. Nucleotide sequence of the cDNA encoding the precursor for mitochondrial serine:pyruvate aminotransferase of rat liver. Eur. J. Biochem. 168:537–542. Page, R. D. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Appl. Biosci. 12:357–358. Purdue, P. E., M. J. Lumb, and C. J. Danpure. 1992. Molecular evolution of alanine:glyoxylate aminotransferase 1 intracellular targeting: analysis of the marmoset and rabbit genes. Eur. J. Biochem. 207:757–766. Springer, M. S., W. J. Murphy, E. Eizirik, and S. J. O’Brien. 2003. Placental mammal diversification and the CretaceousTertiary boundary. Proc. Natl. Acad. Sci. USA 100:1056– 1061. Stewart, C. B., J. W. Schilling, and A. C. Wilson. 1987. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330:401–404. Takada, Y., N. Kaneko, H. Esumi, P. E. Purdue, and C. J. Danpure. 1990. Human peroxisomal L-alanine: glyoxylate aminotransferase: evolutionary loss of a mitochondrial targeting signal by point mutation of the initiation codon. Biochem. J. 268:517–520. Takasuga, A., S. Hirotsune, R. Itoh, A. Jitohzono, H. Suzuki, H. Aso, and Y. Sugimoto. 2001. Establishment of a high throughput EST sequencing system using poly(A) tailremoved, cDNA libraries and determination of 36 000 bovine ESTs. Nucleic Acids Res. 29:e108. Takayama, T., K. Fujita, K. Suzuki, M. Sakaguchi, M. Fujie, E. Nagai, S. Watanabe, A. Ichiyama, and Y. Ogawa. 2003. Control of oxalate formation from L-hydroxyproline in liver mitochondria. Journal of the American Society of Nephrology 14:939–946. Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. von Heijne, G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335–1342. Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS 13:555–556. ———. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15:568–573. Yang, Z., S. Kumar, and M. Nei. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650. Dan Graur, Associate Editor Accepted November 4, 2003