* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PINdb: a database of nuclear protein complexes from human and
Phosphorylation wikipedia , lookup
Cell nucleus wikipedia , lookup
Signal transduction wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Protein folding wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Chemical biology wikipedia , lookup
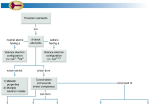
Vol. 20 no. 9 2004, pages 1413–1415 DOI: 10.1093/bioinformatics/bth114 BIOINFORMATICS PINdb: a database of nuclear protein complexes from human and yeast Phuong-Van Luc and Paul Tempst∗ Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10021, USA Received on July 18, 2003; revised on November 18, 2003; accepted on November 25, 2003 Advance Access publication April 15, 2004 ABSTRACT Summary: Proteins Interacting in the Nucleus database (PINdb) is a database of protein complexes purified from the nucleus of human and yeast cells. It is compiled from the published literature and existing databases. Currently, PINdb contains mostly protein complexes that may be involved in gene transcription.To facilitate comparative analyses and identification of protein complexes, the compositional information is integrated with standardized gene nomenclature, annotation and protein sequences from public databases. The PINdb web interface provides a number of tools for (1) comparison of protein complexes, (2) search for a protein complex by its published name or by a partial list of its components and (3) browsing specific subsets or a functional classification of the complexes. Availablity: http://pin.mskcc.org Contact: [email protected] INTRODUCTION Temporal and spatial control of gene expression require a plethora of transcriptional proteins and enzymes. The access of RNA polymerase II to a gene promoter alone is regulated by some 80 plus proteins (Freiman and Tjian, 2003; Narlikar et al., 2002; Orphanides and Reinberg, 2002; Malik and Roeder, 2000), which include general transcription factors, co-activators, co-repressors and chromatin remodelers as well as sequence-specific transcriptional factors. As is the norm for proteins extracted from the cell, those involved in the transcription process often co-purify as large macromolecular complexes, and it is not unusual for some complexes to contain members of other protein assemblies: hence the general belief that proteins exist in the cells as interconnecting or overlapping networks of tightly associated complexes or assemblies. How a protein functions in vivo also appears to be dependent on its macromolecular environment: in uncomplexed forms, many of the purified proteins exhibit either altered or no enzymatic activities. To understand fundamental processes going on in the cell, it is therefore important to map ∗ To whom correspondence should be addressed. Bioinformatics 20(9) © Oxford University Press 2004; all rights reserved. out the physical interactions among the various proteins and protein complexes. Using traditional and laborious methods of biochemical and genetic analysis, investigators have isolated and characterized fewer than 200 cellular and nuclear protein complexes over the past 20 years. However, with the more recent application of mass spectrometry (MS) and computer-aided database searches in purified protein identification, the rate at which new protein complexes are discovered and characterized has accelerated. In fact, innovations in protein complex purification (e.g. affinity capture using FLAG- or TAP-tagged proteins) and MS-based analytical techniques have helped fuel the rapid growth of a new field, proteomics, and made possible large-scale analysis of the proteomes of model organisms (Aebersold and Mann, 2003). The fast pace at which new transcriptional complexes are being described has introduced major challenges for gene regulation and proteomics researchers and especially for those studying human and other mammalian systems. It confounds the ongoing problems of complex identification and definition, as posed by the multitude of names by which a protein or complex is referred, the differences in protein purification methodology and purification stringency, tissue or cell source used and differences in MS analytical approaches used in their identification. To aid researchers in their effort to resolve the identity of protein complexes and associated proteins and to delineate functional roles, we have compiled a literature database of nuclear protein complexes and developed software to integrate this information with gene nomenclature, annotation and sequence information from the HGNC (http:// www.gene.ucl.ac.uk/nomenclature/), LocusLink (http://www. ncbi.nlm.nih.gov/LocusLink/), OMIM (http://www.ncbi.nlm. nih.gov/omim/) and Entrez Protein (http://www.ncbi.nlm.nih. gov/entrez/query.fcgi?db=Protein) databases. A relational schema has been designed to describe the relevant architectural and functional properties of multiprotein factors isolated from nuclear extracts and to facilitate the development of efficient SQL-based tools for comparative analysis of these complexes. Our goal is to provide a specialized knowledge base for proteomics research and to create a framework for the 1413 P.-V.Luc and P.Tempst Fig. 1. Architecture of PINdb. development of a map of physical protein interaction networks and proteomic data mining tools. Currently, there are available in the public domain other databases that archive interacting proteins such as BIND (Bader et al., 2003, http://www.bind.ca/), MIPS (Mewes et al., 2002, http://mips.gsf.de/) and DIP (Xenarios et al., 2002, http://dip.doe-mbi.ucla.edu/). However, Proteins Interacting in the Nucleus database (PINdb) differs from these resources in several aspects. It focuses on multiprotein nuclear complexes from human or yeast cells that have been biochemically and/or functionally characterized. This is in contrast to the comprehensive BIND, which contains protein complexes from various species and cellular compartments and includes unknown complexes reported in high-throughput proteomics studies. To avoid duplication of effort, the yeast portion of PINdb includes a subset of complexes with subunits known to localize in the nucleus extracted from the MIPS Comprehensive Yeast Genome Database (CYGD, http://mips.gsf.de/proj/yeast/CYGD/db/), as well as a subset of interacting protein pairs derived from DIP. These datasets are maintained as separate subdatabases and provide a searchable reference source of yeast nuclear protein interactions for comparison purposes. To the best of our knowledge, none of the publicly accessible knowledge bases provides tools that allow comparative 1414 analysis of multiprotein complexes, subunit name resolution and the capability to browse or search for complexes by cellular functions, by components or by common names. In this respect, PINdb presents a unique resource for the study of protein interations in the nucleus. DATABASE CONTENTS PINdb contains curated information on human and yeast protein complexes that may be found in the cell nucleus. Currently, the human dataset is limited to those pertaining to the RNA polymerase II-dependent transcriptional machinery. The human protein complexes are all derived from the results of biochemical proteomic studies. The yeast subset includes nuclear complexes curated from the published literature as well as those imported from the complexes catalogue of the MIPS CYGD. An overall view of the PINdb system is shown in Figure 1. The information collected for each protein complex includes common names and aliases, methods of isolation, tissue source and compositional, enzymatic and functional properties. We assign a unique ID as well as a unique name (derived from the name given in the publication or its functional role) to each complex to facilitate search and comparison of multiple complexes by name. A classification scheme was developed PINdb to permit efficient retrieval and analysis of PIN complexes by functional roles and by enzymatic properties. Each PIN complex is linked by its unique ID to information regarding subunit proteins, such as names and aliases, apparent and predicted molecular weights, protein sequences and printed references (publications that first describe the proteins as components of the pertinent complexes). Two subdatabases are created to provide a knowledge base for yeast proteins and a reference source for human gene nomenclature. The yeast protein knowledge base integrates pertinent data from CYGD (http://mips.gsf.de/ proj/yeast/CYGD/), SGD (http://genome-www.stanford.edu/ Saccharomyces/) and SWISS-PROT (http://www.ebi.ac.uk/ swissprot/). Detailed information on subunit proteins of curated yeast complexes can be readily retrieved using systematic open reading frame (ORF) gene names. The reference source for human protein names was compiled from the printed literature and from Web databases, such as HGNC, GDB (http://www.gdb.org/gdb/), LocusLink, OMIM and GeneCards (http://bioinformatics.weizmann.ac.il/ cards/). As new protein subunit entries are added to the database, a SQL-based Java utility is periodically executed to search through the curated aliases, find the HGNC-approved name, if one exists, or arbitrarily select an interim name for each protein and use this information to refresh a table storing the official/interim name and all known aliases of each human protein. ARCHITECTURE AND IMPLEMENTATION PIN consists of a relational database back-end (implemented in Microsoft Access on the Windows 2000 platform), a Java application layer (database connection provided by a JDBC driver from NetDirect) and a front-end Apache HTTP server and Tomcat servlet container. Users can access the database over the Internet using their favorite Web browsers. We use servlet and Java server page technology to provide dynamic contents and to enable user interactions with PIN. WEB-BASED TOOLS AND UTILITIES Several SQL-based Java tools are available for content viewing, database search and comparative analysis of protein complexes. Users can browse selected datasets, retrieve complexes by name or by functional category or find all complexes that share some common subunits. The composition of various protein complexes can be compared using Subunit Lineup Tool, which arranges the subunits in adjacent columns of descending molecular weights and places those determined to be common in the same row. Further protein information can be retrieved through hyperlinks to internal data or to comprehensive databases, such as Pubmed, Entrez Protein, LocusLink, SwissProt, SGD and MIPS. Nomenclaturerelated tools include utilities to find a standard ORF name (yeast) or official human gene name and aliases for individual proteins or to retrieve a list of official and common names for all subunits of a protein complex. More details regarding PIN usage and database statistics can be found at http://pin.mskcc.org/ (navigate to About PIN). FUTURE DIRECTIONS We will continue to add new entries and update the PINdb on a frequent basis. The database schema, Web tools and interface will continue to evolve to cope with new findings and developments in gene regulation and proteomics research. REFERENCES Aebersold,R. and Mann,M. (2003) Mass spectrometry-based proteomics. Nature, 422, 198–207. Bader,G.D., Betel,D. and Hogue,C.W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–450. Freiman,R.N. and Tjian,R. (2003) Regulating the regulators: lysine modifications make their mark. Cell, 112, 11–17. Malik,S. and Roeder,R.G. (2000) Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. Mewes,H.W., Frishman,D., Guldener,U., Mannhaupt,G., Mayer,K., Mokrejs,M., Morgenstern,B., Munsterkotter,M., Rudd,S. and Weil,B. (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 30, 31–34. Narlikar,G.J., Fan,H.-Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487 . Orphanides,G. and Reinberg,D. (2002) A unified theory of gene expression. Cell, 108, 439–451. Xenarios,I., Salwinski,L., Duan,X.J., Higney,P., Kim,S.M. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. 1415