* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download BIO00011C Cell and Developmental Biology
Survey
Document related concepts
Hedgehog signaling pathway wikipedia , lookup
Cell nucleus wikipedia , lookup
Protein moonlighting wikipedia , lookup
Cytokinesis wikipedia , lookup
Extracellular matrix wikipedia , lookup
Endomembrane system wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Biochemical switches in the cell cycle wikipedia , lookup
Cellular differentiation wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemical cascade wikipedia , lookup
Transcript
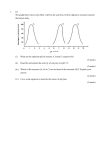
BIO00011C Examination Candidate Number: _____________ Desk Number: _____________ University of York Department of Biology B. Sc Stage 1 Degree Examinations 201516 Cell and Developmental Biology Time allowed: 1 hour and 30 minutes Total marks available for this paper: 50 ● Answer all questions in the spaces provided on the examination paper ● The marks available for each question are indicated on the paper For marker use only: 1 2 3 4 5 6 7 8 9 10 11 For office use only: Total as % page 1 of 13 BIO00011C Answer all questions in the spaces provided 1. What does the flagellar constraint hypothesis suggest about the evolution of cell specialisation? (3 marks) Cell specialisation evolved to overcome the issue of competition for the microtubule organising centre which is required for both cell division and movement of flagella (1 mark). Thus unicellular organisms cannot use their flagella and divide at the same time (1 mark). There was selective pressure for the evolution of flagellate and nonflagellate cells in primitive multicellular organisms, which allowed growth and movement to go on at the same time (1 mark). Module level learning outcome: Understanding the properties of eukaryotic cells that allow the development of multicellular organisms. Feedback: I was a bit disappointed with how this was answered. Most answers got that there was some competition for cellular machinery but the details on the MTOC tended to be vague. To get full marks I wanted some statement about selective pressure favouring evolution of flagellate and nonflagellate cells in primitive multicellular eukaryotes. Some answers explicitly stated that this was going on in prokaryotes, which rather misses the focus of the module. 2. What does the ability to produce an animal clone, like Dolly the sheep, tell us about the regulation of cell differentiation? (3 marks) It demonstrates that genetic information is not lost during the process of cell lineage specification and differentiation (1 mark). Differentiated somatic cells contain all the necessary information to allow normal development (1 mark). Therefore, development is regulated by differential gene expression (1 mark). Module level learning outcome: Understanding the critical role that differential gene expression plays in the life of eukaryotic cells Feedback: Most answers addressed the central issue of gene constancy but a surprising number failed to mention the conclusion that development must therefore be regulated by differential gene expression. page 2 of 13 BIO00011C 3. a) Explain why peptide hormone receptors are usually located at the cell surface. (1 mark) Peptides are typically hydrophilic and soluble in water. They are therefore unable to pass across the hydrophobic cell membrane. Module level learning outcome: Understanding principles of signalling pathways in plants and animals. Feedback: No problems here, most answers were on top of this, but some only got ½ mark because they stated that peptides are generally hydrophilic but failed to mention what this meant in the context of the hydrophobic cell membrane. b) Describe how binding of the FGF ligand to its receptor leads to the activation of the ras protein. (3 marks) Ligand binding causes receptor dimerization and autophosphorylation on intracellular tyrosine residues (1 mark), which allows association of the grb2 adaptor protein with receptor complex (1 mark). A guanine nucleotide exchange factor (GEF) associates with grb2 and causes ras to cycle from its inactive GDP bound form to its active GTP bound form (1 mark). Module level learning outcome: Understanding principles of signalling pathways in plants and animals. Feedback: A spread of answers here. Many nailed the answer but many also lost marks because of lack of precision eg. suggesting that GEF or FGFR phosphorylate ras or ras phosphorylates GDP to GTP and that ras is a kinase. This latter issue is common from year to year despite it being explicitly discussed in the relevant lecture. c) Once activated, how is ras subsequently inactivated? (1 mark) In its active state ras has an intrinsic GTPase activity which hydrolyses GTP to GDP and causes ras to cycle back to its GDP bound inactive state. Module level learning outcome: Understanding principles of signalling page 3 of 13 BIO00011C pathways in plants and animals. Feedback: Many answers correctly identified a GTPase activity as being important but lost ½ mark for not stating that it is intrinsic to ras or implicating that GAP was the GTPase. Again it is an issue with precision. d) Explain how the MAP kinase cascade provides an amplification step in the FGF signal transduction pathway. (3 marks) The components (raf, mek and Erk) of the cascade are all kinase enzymes (1 mark). A single activated raf molecule can phosphorylate and activate multiple downstream MEK molecules. Each activated molecule can then activate multiple Erk molecules. Thus the activation of few raf molecules at the top of cascade leads to the activation of many Erk molecules at the end of the cascade (2 mark). Module level learning outcome: Understanding principles of signalling pathways in plants and animals. Feedback: Quite a lot of answers were quite vague on what constituted the amplification in this pathway. Simply stating the components of the cascade only attracted 1 mark. I wanted some statement regarding one molecule of an enzyme being able activate multiple target molecules. Interestingly some alluded to this idea only in the context of Erk acting on multiple downstream transcription factors and ignoring raf and mek. I has some sympathy for this answer and accurately stated answers of this sort got 1.5 marks. Another reasonably common mistake is the implication that Erk is a transcription factor. This is another example where precision in thinking is called for. 4. a) What is the transcriptional initiation complex and where is it assembled? (2 marks) It is the complex of RNA polymerase II and general transcription factors (1 mark) that is assembled at the promoter of a gene before the start of transcription (1 mark). Module level learning outcome: Understanding the critical role that differential gene expression plays in the life of eukaryotic cells page 4 of 13 BIO00011C Feedback: Mostly answered well but I knocked off ½ for saying transcription factors rather than general transcription factors. b) How are transcription factors bound at distant enhancers able to influence the activity of the transcription initiation complex? (1 mark) Transcription factors bound at enhancers are able to induce bending of the DNA allowing them to contact and influence the activity of the transcription initiation complex at a distant promoter. Module level learning outcome: Understanding the critical role that differential gene expression plays in the life of eukaryotic cells Feedback: Saying that they induced DNA bending or looping was not sufficient to get the full mark. I needed some statement about allowing contact with the transcription initiation complex c) Describe the mechanism by which myogenic regulatory factors regulate the development of skeletal muscle. (3 marks) Myogenic regulatory factors are required for the transcription of genes coding for proteins involved in muscle differentiation e.g. actin, myosin, tropomyosin etc (1 mark). They do this by binding as dimers with Eproteins to pairs of Eboxes in the regulatory regions of the muscle genes (2 marks). Module level learning outcome: How key cellular processes are integrated during the development of tissues and organ systems Feedback: Some students answered this by describing the involvement of the different MRFs at key points in the skeletal muscle differentiation pathway. This was not really what I wanted but if it was accurately described I awarded 1 mark. However, most answers appropriately attempted to described the molecular mechanism of bHLH regulation of gene expression. page 5 of 13 BIO00011C 5. a) How does endocrine signalling differ from paracrine signalling? (1 mark) Endocrine signalling involves long distance systemic transport of the signalling molecule, whereas paracrine signalling involves local signalling over a short distance, perhaps a few cell diameters. Module level learning outcome: Understanding principles of signalling pathways in plants and animals. Feedback: Almost all answers were correct, with the key issue being short distance versus long distance. Depending on how things were phrased I knocked off ½ mark if it wasn’t clear in the answer that systemic transport meant long distance signalling. b) How does a morphogen generate positional information in the developing embryo? (2 marks) A morphogen produced by a source of cells in the embryo generates a gradient (1 mark). Cells are able to sense the concentration of the morphogen within the gradient and determine where they are relative to the source (1 mark). Module level learning outcome: How key cellular processes are integrated during the development of tissues and organ systems Feedback: Most answers mentioned gradient or concentration but to get full marks I wanted some understanding that positional information relative was to some signalling source. Some answers incorrectly implied that morphogens were a type of cell that responded to graded signals, which was disappointing. c) Provide a piece of evidence that TGFbeta/nodal signalling is involved in mesoderm induction. (1 mark) A number of possible answers here: ● Dominant negative activin receptor blocks mesoderm formation ● Treatment of animal cap explants with activin protein induces mesoderm Module level learning outcome: How key cellular processes are integrated during the development of tissues and organ systems page 6 of 13 BIO00011C Feedback: There was quite a lot of imprecision in the way this was answered. I wanted some reference to actual experiments that were discussed in lectures. Simply saying that inhibiting nodal signalling meant that mesoderm induction failed was not sufficient to gain a mark as that could have been guessed from the question. Most correct answers mentioned the experiment using a dominant negative activin receptor, although the details tended to be a bit vague. As long as the general principle was correct I awarded a full mark. Surprisingly few mentioned the biological activity of nodal protein as a mesoderm inducer in animal cap explant experiments. Quite a lot of answers talked about the nodal gradient in the embryo and experiments that involved removing the vegetal hemisphere. This evidence is very indirect and I did not award a mark for these answers. d) How do Smad proteins regulate gene expression? (1 mark) Once phosphorylated and activated they are able to translocate to the nucleus where they act as transcription factors binding to and regulating the transcription of target genes. Module level learning outcome: Understanding principles of signalling pathways in plants and animals. Feedback: Mostly answers well. Some indicated that Smad proteins were kinases. Not every signal transduction component is a kinase! 6. a) In which subcellular compartment are proteins packaged into secretory vesicles? (1 mark) The trans Golgi network or TGN This was generally answered well. It should be noted that secretory vesicles are those formed at the TGN and fuse with the plasma membrane, intraGolgi vesicles (which transport cargo between different Golgi cisternae are not secretory vesicles). Partial marks were awarded for ‘Golgi body/complex/apparatus” b) Name two other organelles that make up the secretory pathway? (2 marks) i) Rough endoplasmic reticulum (ER) (1 mark) page 7 of 13 BIO00011C ii) the Golgi (complex or apparatus) (1 mark) As above this was answered reasonably well. If the answer to a) was given as “The Golgi” and TGN was listed here, then marks were awarded. Marks were also awarded for “Plasma membrane”. Partial marks were awarded for “endosome” as while this is not technically correct there are specialised secretory pathways which encompass endosomes. The most common erroneous answers given were “lysosome” (often misspelt as lyzozyme) which is part of the endosomal pathway, or ribosomes (these are protein/RNA complexes, not organelles). c) Provide one function for each of your answers to part (b). (2 marks) For rough ER either protein folding, quality control or addition of core oligosaccharides (1 mark for either), for Golgi, sequential modification of glycosylation moieties (1 mark). If an incorrect answer (or answers) were given in b) and a function of that cellular component was correctly outlined here then marks were awarded (so as not to penalise students twice). Module level learning outcome: Understanding the properties of eukaryotic cells that allow the development of multicellular organisms. 7. Name an organelle thought to have evolved via endosymbiosis, provide one line of evidence in support of your answer. (2 marks) Either mitochondria or chloroplasts (1 mark). Lines of evidence discussed for these having evolved via endosymbiosis is that they: (1 mark for any) 1. are similar size and morphology to bacteria 2. have membrane composition consistent with absorption of prokaryotic cell by eukaryotic host page 8 of 13 BIO00011C 3. have their own DNA and ribosomes (with distinct characteristics to host), and as in bacteria transcription and translation occur simultaneously in the same compartment. 4. divide by binary fission This was answered well with most students gaining full marks. Module level learning outcome: Understanding the properties of eukaryotic cells that allow the development of multicellular organisms. 8. An experiment was set up to determine changes in protein expression during the cell cycle in embryonic stem cells (ESCs). At 24 hours before the start of the experiment, the ESCs cells were switched to a culture medium without serum to synchronise the cells in G0. The serum was then replaced and protein samples were taken from the ESCs every 2 hours for 14 hours. The protein samples were analysed by SDSPAGE and are shown in the figure below. Figure legend: SDSPAGE analysis of protein expression in ESCs during the cell cycle. After 24 hours of serumstarvation, serum was added back to the cell cultures and protein extracted every 2 hours for 14 hours and loaded onto the gel (lanes 17). The gel was stained to allow visualisation of the different sized proteins. page 9 of 13 BIO00011C a) Describe briefly how you would test the pluripotency of the ESCs. (3 marks) Teratoma assay (1 mark) subcutaneous implantation into nude mice (1) to determine formation of tumours with tissues derived from 3 germ layers (1). Answers may also refer to expression of Oct4, Nanog or Sox2, but a less satisfactory test. Module level learning outcome: Understanding the properties of eukaryotic cells that allow the development of multicellular organisms. Feedback: Most answered this well, getting the key points for full marks. There were several suggestions for in vitro experiments, and credit was given if correct, but not the gold standard teratoma assay. b) The SDSPAGE analysis suggests that the expression levels of one protein clearly changes as the cells progress through the cell cycle. On the figure above, label this protein as “Protein A” with an arrow. (1 mark) See arrow above Module level learning outcome: Understanding the critical role that differential gene (protein) expression plays in the life of eukaryotic cells Feedback: Needed to be accurate and indicate the arrow as above in line with Protein A from all lanes. Singling out an individual band from one lane only is not correct. c) In the cell cycle of ESCs, it is known that M phase lasts about 1 hour, the G1 phase lasts approximately 34 hours, S phase lasts approximately 78 hours and G2 around 46 hours. Referring to the SDSPAGE results, what can you deduce about the expression of Protein A in these ESCs during the cell cycle? (4 marks) At the start of the experiment (serum addition), the cells will enter the cell cycle at G1 (1 mark). In G1 (lane 1), protein A levels are low/absent (1). G1 lasts 34 hours, so lane 2 may represent late G1 or early S phase and Protein A levels begin to increase to reach a peak around 8 hours (lane 4) during S phase (1). S phase will last 78 hours, so from lanes 36 (612 hours after the start of the experiment). After this time, Protein A levels reduce (lane 7) (1). Protein A expression is associated with the S phase of page 10 of 13 BIO00011C the cell cycle (1). Module level learning outcome: Understanding the properties of eukaryotic cells that allow the development of multicellular organisms. Feedback: Answers needed to show an understanding of the experiment and the correct interpretation of the gel, covering G1, S and G2 phases. Answers that only identified an increase in S phase would not get full marks. Errors made in part b) did not influence answers to this question. 9. The schematic below describes the differentiation of a haematopoietic stem cell into an erythrocyte (red blood cell). Suggest names for stages A and B. (2 marks) A: BFUE = BurstForming Unit Erythrocyte. Will also accept proerythroblast, or erythrocyte progenitor. B: CFUE = ColonyForming Unit Erythrocyte. Will also accept erythroblast, or committed erythroblast. Needs to show an understanding of the stagespecific differentiation process and lineage commitment, not just factual recall. Module level learning outcome: How key cellular processes are integrated during the development of tissues and organ systems Feedback: Quite a few answers correctly identified BFUE and CFUE. There were also a range of different suggestions and, as indicated in the model answer, if there were logical demonstrating a staged progression even if the technical terms were not fully correct, full marks were awarded. 10. Give two examples of how endochondral ossification differs from intramembranous ossification during development of the skeleton. page 11 of 13 BIO00011C (2 marks) i) EO involved in the development of long bones, IO in flat bones. ii) EO involves initial differentiation into cartilage, IO direct differentiation into bone. Possible others: ● EO involve marrow cavity formation/haematopoiesis, IO does not. ● EO involved in bone lengthening, IO not. Module level learning outcome: How key cellular processes are integrated during the development of tissues and organ systems Feedback: Impressive number of correct answers. Some marks were lost if IO was not considered in the response, i.e. if only EO was described and not how it differed to IO. Marks also lost if the two suggestion were far too similar to each other. 11. a) Describe the phenotype of flowers from a ‘c’ function mutant in comparison to wildtype. (2 marks) Wildtype flowers have whorls of four floral organs sepals on the outside, followed by petals, stamens and carpels (1 mark). ‘c function’ mutants have a sepals, petals, petals, and sepals (1 mark). LOs: Explain how mutant phenotypes reveal the genes and proteins that regulate flower development MLOs: How key cellular processes are integrated during the development of tissues and organ systems b) Which floral organ develops in a whorl where both ‘a’ and ‘b’ function genes are expressed? (1 mark) The expression of ‘a’ and ‘b’ function genes overlaps in whorl 2 where petals develop. LOs: Describe the ABC model of flower development MLOs: How key cellular processes are integrated during the development page 12 of 13 BIO00011C of tissues and organ systems c) What evidence demonstrates that the activities of ‘a’ and ‘c’ function genes are mutually antagonistic? (3 marks) The ABC model for flower development shows that there are three regulators for organ identity. These functions overlap to generate distinct combinations is each whorl (1 mark). The phenotype of a and c function mutants shows that the ‘a’ and ‘c’ activities must be mutually antagonistic (1 mark). When ‘a’ is missing, the domain of ‘c’ expands and vice versa (1 mark). LOs: Explain how models can be used to test theories that help explain development MLOs: How key cellular processes are integrated during the development of tissues and organ systems page 13 of 13