* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Molecular Biology-restrection enzyme
Silencer (genetics) wikipedia , lookup
DNA barcoding wikipedia , lookup
Gel electrophoresis wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Maurice Wilkins wikipedia , lookup
Non-coding DNA wikipedia , lookup
Agarose gel electrophoresis wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Molecular evolution wikipedia , lookup
SNP genotyping wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Genomic library wikipedia , lookup
DNA supercoil wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Molecular cloning wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
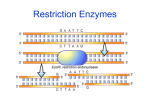
Molecular Biology March 2007 Restriction Enzymes Restriction Enzymes • Restriction enzymes are part of the defense system of bacteria: they digest foreign DNA that enters the bacterial cell. • Each species of bacteria has its own set of restriction enzymes. Each enzyme cuts DNA at a specific short base sequence. For instance, EcoR1 cuts the DNA at the sequence GAATTC, and BamH1 cuts at GGATCC. There are hundreds of restriction enzymes known. • Using properly chosen enzymes, the gene you want can be cut out of the chromosome intact, with very little extra DNA. • Many restriction enzymes give a staggered cut across the DNA double helix. This produces short single stranded regions, called “sticky ends”. The ends are sticky because they spontaneously pair with similar ends. Restriction Endonucleases z Restriction endonucleases cut DNA z Present in bacteria z Cut at sequence specific sites – Usually 4 or 6 base pairs long z Bacteria use them to destroy ‘foreign’ DNA – Bacteria protect their own DNA against self-cutting by special methylation of their DNA z Restriction enzymes can be purified and are used in genetic engineering studies Restriction Endonucleases • Example Restriction enzymes – EcoR I (E. coli Restriction Endonuclease I) – Stu I (Streptomyces tubercidicus I) EcoR I Palindromic Axis of rotational symmetry 5’ 3’ GAATTC 3’ 5’ CTTAAG Sticky Ended Stu I 5’ 3’ AGGCCT 3’ 5’ TCCGGA Blunt Ended Molecular Scissors and Glue zThere are 200’s of restriction enzymes, each one with a different recognition site – These enzymes are ‘molecular scissors’ and can be used to specifically cut long DNA strands into smaller pieces Restriction Enzymes ¾ called "restriction enzymes“ because restrict host range for certain bacteriophage ¾ bacterial" immune system": destroy any "non-self" DNA ¾ methylase recognizes same sequence in host DNA and protects it by methylating it; restriction enzyme destroys unprotected = non-self DNA (restriction/modification systems) Restriction Enzymes ¾ Most recognize and cut palindromic sequences ¾ Many leave staggered (sticky) ends ¾ by choosing correct enzymes can cut DNA very precisely ¾ Important for molecular biologists because restriction enzymes create unpaired "sticky ends" which anneal with any complementary sequence ¾ Hundreds of restriction enzymes have been identified. Some Commonly Used Restriction Enzymes Eco RI 5'-G | AATTC Eco RV 5'-GAT | ATC Hin D III 5'-A | AGCTT Sac I 5'-GAGCT | C Sma I 5'-CCC | GGG Xma I 5'-C | CCGGG Bam HI I 5'-G | GATCC Pst I I 5'-CTGCA | G Theoretical Basis Using Restriction Enzymes ¾ The activity of restriction enzymes is dependent upon precise environmental condtions: PH Temperature Salt Concentration Ions ¾ An Enzymatic Unit (u) is defined as the amount of enzyme required to digest 1 ug of DNA under optimal conditions: 3-5 u/ug of genomic DNA 1 u/ug of plasmid DNA Stocks typically at 10 u/ul Agarose gel electrophoresis of the PCR products for the 3'UTR of the leptin receptor gene. Lane 1: a 100 bp ladder, lanes from 2 to 20: the amplified fragments of the 3'UTR region. A PCR assay for the CTTTA insertion at the 3'UTR of the leptin receptor gene A PCR assay for the CTTTA insertion at the 3'UTR of the leptin receptor gene. A fragment of 114 bp or 119 bp in size was amplified, digested with the enzyme Rsal and analyzed by agarose gel electrophoresis, Lane 1: molecular size marker (a 100 bp ladder); lane 2,7and 10: undigested sample, lanes 3, 4, 5, 6 and 9: individuals with genotypes heterozygous( del /ins, - + ), lanes 6 and 8: individuals with genotypes homozygous ( ins/ins, + + ). Thank you