* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture#20 - Gene Interactions and Epistasis
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic drift wikipedia , lookup
X-inactivation wikipedia , lookup
Minimal genome wikipedia , lookup
Gene desert wikipedia , lookup
Behavioural genetics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genetic engineering wikipedia , lookup
Heritability of IQ wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Ridge (biology) wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Human genetic variation wikipedia , lookup
Genome evolution wikipedia , lookup
Public health genomics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genomic imprinting wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Population genetics wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
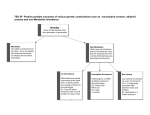
6/24/17 BIOLOGY 207 - Dr. Locke Lecture#20 - Gene Interactions and Epistasis Required readings and problems: Reading: Open Genetics, Chapter 6.2 Problems: Chapter 6 Optional Griffiths (2008) 9th Ed. Readings: pp 239-248 Problems: 9th Ed. Ch. 16: 26-48, 54-66 Campbell (2008) 8th Ed. Readings: Concept 14.3 Concepts: How do alleles at different gene loci interact in diploids? 1. Phenotypic expression of some alleles varies due to "genetic background" or other causes (penetrance, epistasis, genetic suppression). 2. The interaction of genes can modify the typical Mendelian ratios of phenotypic classes of the progeny. 3. Penetrance and expressivity measure frequency and intensity (respectively) of phenotypic expression of a particular genotype. Biol207 Dr. Locke section Lecture#20 Fall'11 page 1 6/24/17 Genetic Background The phenotypic expression (mutant phenotype) of a mutant allele can be modified by different alleles at other gene loci. Why do some characters vary in their expression? 1. Biological systems are complex -> slight differences in the expression of one or more genes in the system -> variation in mutant phenotype 2. Background effects can be due to one, a few, or due to many genes. 3. Phenotypic expression that depend upon the combined effect of a large number of genes is called the genetic background -> additive effect of all the allelic differences in the remainder of the genome. Example of one gene: The case of Biol207 Dr. Locke section recessive epistasis. Lecture#20 Fall'11 page 2 6/24/17 Phenomenon of Recessive Epistasis Recessive epistasis occurs when the phenotypic expression of genotypes at one locus depends on the alleles at another locus. Recessive epistasis can alter phenotypic ratios. E.g. - not 9:3:3:1 (F2 cross) or not 1:1:1:1 (test cross). Example of recessive epistasis: “C” gene in mouse coat colour There are many loci that affect coat colour in mice. B/b locus B = Black pigment b = brown pigment C dominant = permits coat colour expression (of other gene affecting coat colour) c recessive = prevents coat colour expression cc, homozygous recessive lack any coat pigmentation - called albinos - white hair Because they have hair with no pigment, cc prevents (precludes) the expression of any other hair colour loci Labrador retrievers coat colour B/b locus B = Black pigment b = brown pigment E/e locus E = permit deposition e = prevent deposition Biol207 Dr. Locke section Lecture#20 Fall'11 page 3 6/24/17 Example Crosses: Mice: Interaction between the B locus and C locus in (albino) (brown) BBcc x bbCC P1 BbCc (Black) F1 Cross F1 x F1 F2:ratios modified ratio B_C_ black 9 bbC_ brown 3 B_cc albino-white bbcc albino-white 4 Result: The cc homozygote is said to be epistatic to the other coat colour genes The albino phenotype prevents one from "seeing" the phenotype from the other loci Dogs: Interaction between the B locus and E locus in (yellow) (brown) BBee x bbEE P1 BbEe (Black) F1 Cross F1 x F1 F2:ratios modified ratio B_E_ black 9 bbE_ chocolate 3 B_ee yellow bbee yellow 4 Result: The ee homozygote is said to be epistatic to the other coat colour genes The yellow phenotype prevents one from "seeing" the phenotype from the other loci Opposite of epistatic is hypostatic Bb locus is hypostatic to the cc alleles. Biol207 Dr. Locke section Lecture#20 Fall'11 page 4 6/24/17 Modified Phenotypic Ratios produced by 2 gene interactions. Ratio: Type genotype: None 9 3 3 1 A-B- A-bb aaB- aabb Ratio 9 3 3 1 9:3:3:1 AB Ab aB ab Recessive epistasis 9 3 4 9:3:4 of aa acting on B and b alleles AB b a Dominant epistasis 12 3 1 12:3:1 of A acting on B and b alleles A aB ab Duplicate genes 15 1 15:1 A a Complementary genes 9 7 9:7 A a Recessive suppression 9 3 4 13:3 by aa acting on bb B b B Dominant suppression 15 1 15:1 by A acting on bb B b Shading represents combined classes. See extra page on modified Mendelian ratios: http://www.biology.ualberta.ca/courses.hp/bio207.hp/locke/modified_ratios.htm Biol207 Dr. Locke section Lecture#20 Fall'11 page 5 6/24/17 Genetic suppression Suppressor mutation has the effect of suppressing (reducing) the phenotypic expression of a mutation -> results in a more wild type (less mutant) phenotype. Can be: - intra-genic (same allele) - similar to a revertant - extra-genic (different gene locus) Dominant Suppression Example: white-mottled allele (wm) gives mosaic expression of white wm / wm = white mottled phenotype wm / w+ = wm / w- = Dominant Suppressors of wm : wm / wm ; Su- / + or wm / wm ; Su- / Su- Biol207 Dr. Locke section = w+ phenotype Lecture#20 Fall'11 page 6 6/24/17 Cross: w+/w+; Su-/Su- X wm/wm ; Su+/ Su+ P1 w+/wm; Su/Su+ w+ / _ wm/ wm w+ / _ wm/ wm ; Su- / _ ; Su- / _ ; Su+ / Su+ ; Su+ / Su+ F1 9 3 3 1 red red red mottled 15 F2 1 Note: similar ratios to gene duplication (also 15:1 ratio) Enhancer mutations Opposite of suppressors -> they make the phenotype more mutant (enhance the mutant phenotype). Many Genes -> genetic background A gene acts in conjunction with other genes and with the environment. Biol207 Dr. Locke section Lecture#20 Fall'11 page 7 6/24/17 Penetrance & Expressivity Penetrance - Is it there or not? Penetrance - The percentage of individuals with a given genotype in a population who exhibit the phenotype associated with that genotype Note: It may be that not all individuals with genetype a/a exhibit the a phenotype because of suppressors, epistatic genes etc., environment Example: In my lab -> a strain that shows incomplete penetrance of an eye "bleb" phenotype Expressivity - It's there, but how strong is it? Expressivity - the variation in the extent to which a given genotype is expressed phenotypically in a individual or population From my lab: Drosophila wing mutant has a variably expressive phenotype - interruption of a vein in the Drosophila wing - variable loss of the vein There is fly-to-fly variation in the extent to which this character is expressed A range of expression in the population. complete 20% Biol207 Dr. Locke section partial 40% little 30% no loss 10% Lecture#20 Fall'11 page 8 6/24/17 Wild type incomplete penetrance Variable expressivity mutant complete penetrance constant expressivity Variable penetrance and expressivity. Biol207 Dr. Locke section Lecture#20 Fall'11 page 9