* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lect13_HistonesChromatin
List of types of proteins wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcription factor wikipedia , lookup
Genomic imprinting wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Gene expression profiling wikipedia , lookup
Ridge (biology) wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Genome evolution wikipedia , lookup
Gene regulatory network wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Epitranscriptome wikipedia , lookup
Ligand binding assay wikipedia , lookup
Cooperative binding wikipedia , lookup
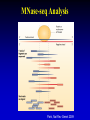
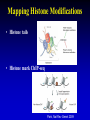
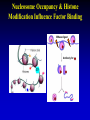
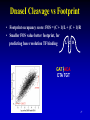
Epigenetics Continued Xiaole Shirley Liu STAT115, STAT215, BIO298, BIST520 Components • DNA-methylation • Nucleosome position • Histone modifications • Chromatin accessibility • Higher order chromatin interactions • Analogy Nucleosome Occupancy & Histone Modification Influence Factor Binding MNase-seq Zentner & Henikoff, Nat Rev Genet 2014 MNase-seq Analysis Park, Nat Rev Genet 2009 MNase-seq Analysis Park, Nat Rev Genet 2009 Factors Influencing Nucleosome Positioning • Rotational positioning from sequence • Statistical positioning from trans-factors (e.g. TF binding) Break Jiang & Pugh, Nat Rev Genet 2009 Components • DNA-methylation • Nucleosome position • Histone modifications • Chromatin accessibility • Higher order chromatin interactions • Analogy Mapping Histone Modifications • Histone tails • Histone mark ChIP-seq Park, Nat Rev Genet 2009 Histone Modifications • Different modifications at different locations by different enzymes Histone Modifications in Relation to Gene Transcription Bisulfite-Seq H3K27ac H3K4me1 H3K4me3 H3K36me3 H3K27me3 H3K9me3 RefSeq genes SRPK1 SLC26A8 MAPK14 From Ting Wang, Wash U Histone Modifications • Gene body mark: H3K36me3, H3K79me3 • Active promoter (TSS) mark: H3K4me3 • Active enhancer (TF binding) mark: H3K4me1, H3K27ac • Both enhancers and promoters: H3K4me2, H3/H4ac, H2AZ • Repressive mark: H3K27me3, H3K9me3 Annotate / segment the genome based on histone marks 12 13 lncRNA Identification • H3K4me3 active promoters • H3K36me3 transcription elongation Guttman et al, Nat 2009 14 Nucleosome Occupancy & Histone Modification Influence Factor Binding MNase digest Antibody for TF Combine Tags From All ChIP-Seq Extend Tags 3’ to 146 nt Check Tag Count Across Genome Take the middle 73 nt Use H3K4me2 / H3K27ac Nucleosome Dynamics to Infer TF Binding Events /ac /ac /ac /ac /ac Condition 1 Condition 2 Nucleosome Stabilization-Destabilization (NSD) Score He et al, Nat Genet, 2010; Meyer et al, Bioinfo 2011 19 Condition-Specific Binding, Epigenetics and Gene Expression C1 C1 C2 C2 • Condition-specific TF bindings are associated with epigenetic signatures • Can we use the epigenetic profile and TF motif analysis to simultaneous guess the binding of many TFs together? Genes TF1 TF2 TF3 Epigenetics 20 Predict Driving TFs and Bindings for Gut Differentiation 21 Identify Major TF Modules Regulating Gut Differentiation and Function GATA6 Cdx2 Embryonic and organ development genes Cdx2 HNF4 Metabolic and digestive genes • Nucleosome dynamics now applied to hematopoiesis and cancer cell reprogramming Break 22 Verzi et al, Dev Cell, 2010 Components • DNA-methylation • Nucleosome position and histone modifications • Chromatin accessibility • Higher order chromatin interactions • Analogy DNase Hypersensitive (HS) Mapping • DNase randomly cuts genome (more often in open chromatin region) • Select short fragments (two nearby cuts) to sequence • Map to active promoters and enhancers Ling et al, MCB 2010 DHS Peaks Capture Most TF Binding Sites • Motif occurrence in the DHS peaks suggest TF binding • Quantitative signal strength also suggest binding stability Thurman et al, Nat 2012 TF Network from DNase Footprint 26 DnaseI Cleavage vs Footprint • Footprint occupancy score: FOS = (C + 1)/L + (C + 1)/R • Smaller FOS value better footprint, for L C R predicting base resolution TF binding GAT ACA CTA TGT 27 DnaseI Cleavage vs Footprint • Footprint occupancy score: FOS = (C + 1)/L + (C + 1)/R • Smaller FOS value better footprint, for L C R predicting base resolution TF binding • Intrinsic DNase cutting bias could have 300-fold difference, creating fake footprints GAT ACA CTA TGT CAGATA CAGATC … ACTTAC ACTTGT 0.004 0.004 1.225 1.273 28 Using DNaseI “Footprint” to Predict TF Binding • Using base-pair resolution cleavage pattern (“footprint”) hurts TF binding prediction when it is similar to intrinsic DNaseI cutting bias 29 Using DNaseI “Footprint” to Predict Novel TF Motifs Break 30 He et al, Nat Meth 2013 Components • DNA-methylation • Nucleosome position and histone modifications • Chromatin accessibility • Higher order chromatin interactions • Analogy HiC • In situ HiC has excellent resolution • Domains conserved between cells and even species 32 HiC • Loops are condition-specific – Assign binding to genes • Convergent CTCF at domain anchors – CTCF as insulators Epigenetics and Chromatin 34 Transcription and Epigenetic Regulation • Stem cell differentiation • Aging brain • Cancer