* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Poster
Homology modeling wikipedia , lookup
Protein domain wikipedia , lookup
Protein folding wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein purification wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
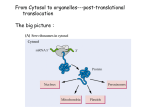
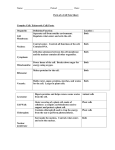
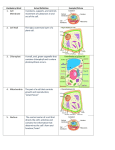
Transportin’ With Transportin A Nuclear Import Mechanism Westosha Central SMART Team: Julia Alberth, Jonah Arbet, Nick Bielski, Monica Ceisel, Sam Colletti, Evan Kirsch, Mitchell Kirsch, Sean Quist, A.J. Reeves and Julia Williams Teacher: Jonathan Kao Mentor: Mark T. McNally, Ph.D., Medical College of Wisconsin I. Abstract III. Transportin’s Anatomy Proteins manufactured in the cytoplasm play an important role in nuclear processes such as RNA splicing. Immediately after transcription, precursor (pre-) mRNA contains introns that are removed in making mature mRNA. Splicing proteins like hnRNP A1 (A1), manufactured in the cytoplasm, are transported into the nucleus and influence RNA splicing decisions. Some proteins in eukaryotic cells use the receptor Transportin (Trn1) for import. Cytoplasmic Trn1 is found in a configuration that allows for the pick-up of cargo proteins. A1 has a nuclear localization signal (NLS) to which Trn1 can bind. Once bound, the Trn1/A1 complex enters the nucleus through a nuclear pore. The protein Ran, when associated with GTP, binds to the complex and causes a loop of approximately 60 amino acids to move and expel the cargo. With the cargo delivered, Trn1 returns to the cytoplasm as a Trn1/RanGTP complex. GTP is then hydrolyzed into GDP signaling Ran to release Trn1. The amino acid loop returns to its original position allowing for another round of transport. The model constructed using 3D printing technology by the Westosha Central HS Smart team in cooperation with MSOE features Trn1 in complex with RanGTP, the mechanical state of cargo unloading. The NLS of A1 can be modified such that Trn1 cannot bind and deliver A1 to the nucleus. Failure of A1 to reach the nucleus results in altered splicing of mRNA, which can lead to diseases like cancer. Therefore, targeting the interaction between Trn1 and its cargo may provide an option for treating diseases. V. NLS Rules Ran Protein Amino Acid Loop Free floating protein in the nucleus that expels cargo from Trn1. This loop of amino acids expel cargo proteins based on the state of RAN. “Cargo” Binding Site This is the region where the NLS binds: it is obscured in this model. Based on 1QBK.pdb Fig. 3 The Transportin Protein2 II. Introduction The central dogma of genetics states DNA is transcribed into RNA which is translated into a protein. Transcription, the formation of RNA coded by a strand of DNA, occurs in the nucleus while translation, the formation of proteins coded by RNA, occurs in the cytoplasm. During transcription, RNA polymerase reads a a gene and transcribes DNA into mRNA. Before the mRNA departs the nucleus, it almost always modified via splicing (See Fig. 1). Fig. 5. Results of the pull down assay5. The left lane of each gel panel shows the results of pulldowns with Trn1 and a predicted NLS. The right lanes show the results including RanGTP, which is known to disrupt Trn1/cargo interactions. IV. Nuclear Import Cycle 1. 2. 3. 4. 5. 6. Pre-mRNA Intron Exon Transportin recognizes the NLS and binds to the cargo protein . Transportin carries the protein through a nuclear pore and into the nucleus. RanGTP binds to Trn1 causing an amino acid loop to expel the cargo. Transportin/Ran complex returns to the cytoplasm. GTP is hydrolyzed to GDP causing Ran to release from transportin. The amino acid loop returns to its original position and transportin can bind to new cargo. Exon Exon 1. Exon Cargo Exon Exon Mature mRNA Transportin + In the process of transcription, the entire gene is read. The resultant pre-mRNA may contain sections that are not represented in the final mRNA. Splicing is the process whereby these unwanted sections of RNA, introns, are removed and the remaining RNA section (exons) are fused together to create a mature mRNA. Alternative splicing is the process where exons are spliced in different combinations, allowing one gene to code for many different proteins. hnRNP A1 (A1) is a protein that influences alternative splicing decisions. If A1 fails to be imported to the nucleus, alternative splicing is changed and, consequently, the protein repertoire of the cell is changed. A1, like all proteins, is manufactured in the cytoplasm. It must be transported into the nucleus to carry out its function. One mechanism by which nuclear import is accomplished uses the protein Transportin (Trn1). Trn1 recognizes a Nuclear Localization Signal (NLS) contained within some proteins that are destined for the nucleus. Trn1 binds to the cargo protein and carries it to the nucleus through a nuclear pore. Cytoplasm 6. Nucleus 2. 2. VI. Biological Significance Ran protein Fig. 6 Modification of a NLS can lead to impaired nuclear import. Phosphorylation of the NLS reduces the affinity between cargo and the transport protein. As a result, the cargo does not bind and reach the nucleus. Figure adapted from Jans et al., 2008. 4. 4. 5. hnRNP A1 In the case of A1 cargo, it influences alternative splicing of many RNAs Conclusion: •Solving the structure of Trn1 allows for prediction of sequences that interact with the cargo binding site. Nuclear pore 3.3. release Results: •GST was always pulled down (linked to beads) •GST alone does not bind to Trn1 (negative control) •All predicted NLSs bind to Trn1 (first lane of each panel) •RanGTP prevents interaction of NLS and Trn1 (second lane of each panel) Splicing is regulated by proteins that need to get to the nucleus. If they fail to get to the nucleus, regulation of splicing goes awry1. The amino acid loop of Trn1 allows deposition of cargo molecules, like splicing proteins, into the nucleus when RanGTP is bound. The NLS of A1 can be modified (see Fig. 6) such that it will not bind to Trn1. Intron removed Fig. 1 The Central Dogma of Genetics & Splicing7 The sequence of the NLS recognized by Trn1 varies from protein to protein. The scientists Lee et al. attempted to predict NLS sequences that interact with transportin using the determined crystal structure of Trn1. Predicted NLSs were attached to a micro-bead as fusion proteins with GST, a protein used to attach the NLS to the micro-bead, then run through a pull down assay and gel electrophoresis. Fig. 4 The Nuclear Import Pathway The SMART Team Program (Students Modeling A Research Topic) is funded by a grant from NIH-SEPA 1R25OD010505-01 from NIH-CTSA UL1RR031973. Failure of A1 to reach the nucleus can cause changes in alternative RNA splicing and the production of inappropriate proteins. These proteins have the potential to cause deleterious conditions including cancer3,6. References 1. Caceres et al. (2000). The MKK3/6-p38–signaling Cascade Alters the Subcellular Distribution of hnRNP A1 and Modulates Alternative Splicing Regulation. The Journal of Cell Biology 149:307-316 2. Chook, M., Blobel, G. (1999). Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 399:230-237 3. David, C.J., Manley, J.L. (2010). Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24:2343-2364 4. Jans et al. (2008). Regulated nucleocytoplasmic trafficking of viral gene products: A therapeutic target? BBA – Proteins and Proteomics 1784:213-227 5. Lee et al. (2006). Rules for the nuclear locatlization sequence recognition by karyopherin beta 2. Cell 126:543-558 6. Lopez-Bigas, N., Audit, B., Ouzounis, C., Parra, G., Guigo, R. (2005). Are splicing mutations the most frequent cause of hereditary disease? FEBS Letters 579:1900-1903 7. http://plantphys.info/plant_physiology/enzymebasics.shtml