* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download ovo D1
History of genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Behavioural genetics wikipedia , lookup
Genome evolution wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Frameshift mutation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Oncogenomics wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression profiling wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression programming wikipedia , lookup
Genome (book) wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Koinophilia wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Genetic drift wikipedia , lookup
Population genetics wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Mutations (changes in DNA):
the lifeblood of genetic analysis
fact: it’s easier to mess things up than to make them better
hence most mutations
with any functional consequence (mutant phenotype)
disrupt normal (wildtype) gene function
Mutations (changes in DNA):
the lifeblood of genetic analysis
Geneticists make mutations
that disrupt normal gene functions
(thereby creating a functional difference between alleles)
to discover what genes do what
…so they can tell biochemists where to look
to learn how those genes do what they do
(…and sometimes geneticists can learn a thing or two
about “how” even before biochemists enter the picture)
Mutations (changes in DNA):
the lifeblood of genetic analysis
Not all mutant phenotypes are equally informative.
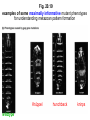
Fig. 20.19
examples of some maximally informative mutant phenotypes
for understanding metazoan pattern formation
Krüppel
wildtype
hunchback
knirps
Forward genetics:
start with
mutant phenotype
ultimately determine
wildtype molecules
(functional consequences
of disruption)
to infer wildtype gene function
(learn molecular mechanism)
Reverse genetics:
start with
wildtype molecules
(ultimately transcription units
a.k.a. genes)
ultimately determine
mutant phenotype
(functional consequences
of disruption)
to infer wildtype function
(…and learn molecular mechanism)
Mutations:
the lifeblood of genetic analysis
…to infer wildtype gene functions
Not Mendel’s goal:
Not Morgan’s goal:
(predict the appearance and
breeding behavior of hybrids)
(learn how inherited information is
transmitted & how it changes)
(neither had any hope of discovering what gene are)
“What are the genes? What is the nature of
the elements of heredity that Mendel
postulated as purely theoretical units? …
Frankly, these are questions with which
the working geneticist has not much
concern himself…
T.H. Morgan
The Relation of Genetics
to Physiology and Medicine
Nobel Lecture, June 4, 1934
Mutations:
the lifeblood of genetic analysis
a Morgan "student":
(1) What kinds can we make? (functional categories)
Herman Muller (1930s):
inferred how mutations can affect gene functioning.
(2) How do we make them? (mutagenesis)
Muller: spontaneous & radiation-induced
(3) How do we find them? (mutant screens & selections)
Muller: exploited giant polytene chromosomes
& invented balancer chromosomes
(Nobel
Prize
1946)
Muller categorized mutations with respect to
change in gene function relative to wildtype
Loss-of-(wildtype)function (l-o-f) mutant alleles
generally recessive (a+/a-: one functional copy “suffices”)
(1) so long as all other genes ok)
(2) so long as we don’t look too hard
clear exception: l-o-f mutations in haploinsufficient genes
are dominant by definition (Minute mutations in flies; Df(M)/+=M/+)
lof alleles causing cancer: identify tumor suppressor genes
“Gain”-of-(over wildtype)function (g-o-f) mutant alleles
generally dominant (often misexpression, wrong time/place): AntpX/Antp+
fly leg where antenna should be
gof alleles causing cancer: identify proto-oncogenes
Loss-of-(wildtype)function (l-o-f) mutant alleles
complete lof: amorph(ic) (null)
Important goal of genetic analysis:
define the null phenotype
partial lof:
hypomorph(ic) (leaky)
phenotypic series: (helpful with pleiotropy)
set of alleles with progressively less function
“Gain”-of-(over wildtype)function (g-o-f) mutant alleles
too much of a good thing:
hypermorph(ic)
something new & different:
neomorph(ic):
different in kind (e.g. fusion protein)
wrong time/place: ectopic expression
antagonizes (poisons)wildtype: antimorph(ic)
(dominant negative)
Muller did not just define the five basic ways the
functioning of a gene can be changed by mutation
(without, he noted, changing its ability to faithfully replicate)
He gave us operational tests to determine
to which class a given mutant allele might belong
Loss-of-(wildtype)function (l-o-f) mutant alleles
complete lof: amorph(ic) (null)
partial lof:
hypomorph(ic) (leaky)
“Gain”-of-(over wildtype)function (g-o-f) mutant alleles
too much of a good thing:
hypermorph(ic)
something new & different:
neomorph(ic):
antagonizes (poisons)wildtype: antimorph(ic)
Muller’s tests: how does the phenotype change when you:
(1) hold the number of mutant alleles constant
and change the number of wildtype alleles.
(2) hold the number of wildtype alleles constant
and change the number of mutant alleles.
increased dose of mutant, phenotype more wildtype
the white gene started it all, and has kept it all going to this day
w+
wapricot
w-
wa/wa (darker, more “wildtype”) > wa/Df(w)
1 copy wa allele
2 copies wa allele
wa/Y (lighter, less “wildtype”) < wa/Y; Dp(wa)/+
1 copy wa allele
2 copies wa allele
See how the phenotype changes when you:
(1) hold the number of mutant alleles constant
and change the number of wildtype alleles.
increased dose of wildtype, phenotype more wildtype
(2) hold the number of wildtype alleles constant
and change the number of mutant alleles.
increased dose of mutant, phenotype more wildtype
wa/w+ (darker, more “wildtype”) > wa/Df(w)
0 copies w+ allele
1 copy w+ allele
wa/Y (lighter, less “wildtype”) < wa/Y; Dp(w+)/+
0 copies w+ allele
1 copy w+ allele
Loss-of-(wildtype)function (l-o-f) mutant alleles
complete lof: amorph(ic) (null)
partial lof:
hypomorph(ic) (leaky)
wapricot
“Gain”-of-(over wildtype)function (g-o-f) mutant alleles
too much of a good thing:
hypermorph(ic)
something new & different:
neomorph(ic):
antagonizes (poisons)wildtype: antimorph(ic)
wapricot
partial lof:
hypomorph(ic)
Increased dose of mutant: phenotype more wildtype
Increased dose of wildtype: phenotype more wildtype
But isn’t in RATHER curious then that:
wa/wa = (same color as) wa/Y
O
+
Muller thought so,
and realized that he had discovered
X-chromosome dosage compensation:
XX=XY (more about that later)
O
complete lof: amorph(ic)
w-
Increased dose of mutant: No change in phenotype
Increased dose of wildtype: phenotype more wildtype
partial lof:
hypomorph(ic)
wapricot
Increased dose of mutant: phenotype more wildtype
Increased dose of wildtype: phenotype more wildtype
Truth be known, geneticists take shortcuts (c.f. cis/trans test)
The characterization of a mutant allele as
a amorphic (null) vs. hypomorphic
is generally made based only on a comparison of
the homozygous mutant to the hemizygous mutant
(and with the knowledge that the mutant is recessive):
whiteX/whiteX vs. whiteX/Df(w)
…potential pitfalls, but a good place to start
“Gain”-of-(over wildtype)function (g-o-f) mutant alleles
too much of a good thing:
hypermorph(ic)
Increased dose of mutant: phenotype more mutant
Increased dose of wildtype: phenotype more mutant
something new & different:
neomorph(ic):
Increased dose of mutant: No change or more mutant in phenotype
Increased dose of wildtype: No change in phenotype
our friend from the X-files, Antp
antagonizes (poisons)wildtype: antimorph(ic)
Increased dose of mutant: phenotype more mutant (if possible)
Increased dose of wildtype: phenotype more wildtype
ovoD1
to infer the normal function of a gene:
LOF alleles simplest to interpret
GOF alleles usually more interesting
but generally harder to interpret
.. especially if don’t know amorph (null) phene.
consider the fly gene ovo as a good example of the problems that arise:
ovoD(ominant)#1 dominant female-specific sterile antimorph
ovoe8K recessive female-specific sterile hypomorph
Both X-linked & similar phenotypes, but can't directly map ovoD1
How can we determine whether ovoD1 and ovoe8K are alleles?
complementation test? (ovoD1/ovoe8K?)
How can we determine whether ovoD1 and ovoe8K are alleles?
Can use GOF alleles to generate LOF alleles relatively easily:
“revert” (suppress) the dominance of a GOF allele
LOF allele
(make it stop doing something bad by inducing LOF mutation IN CIS)
…then establish allelism using the GOF-LOF double mutant alleles
-- also helps define the null phenotype
GOF
{
ovoD1 / +
Df(ovo) / +
tumorous female germline (dominant)
not haploinsufficient wildtype
}
female germline
LOF
ovoe8K / +
wildtype female germline
ovoe8K / ovoe8K
tumorous female germline (recessive)
can't use the complementation test:
ovoD1 / ovoe8K
wildtype or mutant?
neither would tell us anything
cis-trans test?
…if in same gene, cis phenotype ≠ trans
ovoD1 + / + ovoe8K
ovoD1 ovoe8K / + +
tumorous
less tumorous or wt.
…if in different genes, cis phenotype = trans
ovoD1 + / + ovoe8K
ovoD1 ovoe8K / + +
tumorous
tumorous
…if in same gene, cis phenotype ≠ trans
ovoD1 + / + ovoe8K
ovoD1 ovoe8K / + +
tumorous
less tumorous or wildtype
but how construct?
Consider a slightly different cis/trans situation:
( ovoD1 + / + +
ovoD1 + / + ovo-null
ovoD1 ovo-null / + +
tumorous )
tumorous
wildtype
Consider a slightly different cis/trans situation:
tumorous
wildtype
ovoD1 + / + ovo-null
ovoD1 ovo-null / + +
what we have:
what we want to generate:
ovoD1 + / + +
ovoD1 ovo-null / + +
but what if true reversion: + +/+ + ?
ovoD1 ovo-null
tumorous
mutagenize to
“revert”
the dominance
wildtype
look at homozygote
/
ovoD1 ovo-null
double mutant homozygote will be tumorous
ovoD1 + / + +
what we have:
what we want to generate:
ovoD1 ovo-null / + +
now do complementation test
to distinguish the alternatives:
tumorous
mutagenize to
“revert”
the dominance
wildtype
ovoD1 ovo-null / + ovoe8K
tumorous
/ + ovoe8K
wildtype
vs.
++
ovoD1 + / + +
ovoD1 ovo-null / + +
tumorous
mutagenize to
“revert”
the dominance
wildtype
…allowed us to do a complementation test:
ovoD1 ovo-null / + ovoe8K
tumorous
/ + ovoe8K
wildtype
vs.
++
Failure to complement with ovoe8K means that
we have to lose the gene function that ovoe8K is missing
in order to “revert” (suppress) the dominance of ovoD1.
..hence we have established that
ovoD1 and ovoe8K are functional alleles
Same for Antp32a5 and Ns1, two GOF mutant alleles
that cause the antenna to develop as a leg instead.
revert their dominance
(one gets a recessive lethal in each case)
…the resulting recessive lethal mutant chromosomes
fail to complement.
(I owe my own success to reverting dominant mutations)