* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Noushin Farnoud Presentation
Oncogenomics wikipedia , lookup
Essential gene wikipedia , lookup
Gene expression programming wikipedia , lookup
Genomic imprinting wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Microevolution wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome (book) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Designer baby wikipedia , lookup
Minimal genome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression profiling wikipedia , lookup
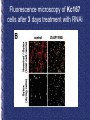
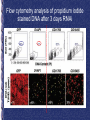
Genome-Wide RNAi Analysis of Growth and Viability in Drosophila Cells Michael Boutros, Amy A. Kiger, Susan Armknecht, Kim Kerr, Marc Hild, Britta Koch, Stefan A. Haas, Heidelberg Fly Array Consortium, Renato Paro, Norbert Perrimon Presented by: Noushin Farnoud Goal Whole Genome Sequence Using RNAi to analyze LOF phenotypes to characterize gene functions List the putative Genes in cell growth and viability Functional Analysis of predicted Genes Cell viability assay: Using Lucimnescent to correlate the reduction of signal to dying cells • Fluorometric method for estimating the number of viable cells in culture by quantifying the amount of ATP present. inhibitor of apoptosis : positive control in screen Fluorescence microscopy of Kc167 cells after 3 days treatment with RNAi Z scores • Indicates how far and in what direction an item deviates from its distribution's mean. • Especially useful when seeking to compare the relative items from distributions with different means and/or different standard deviations. Method Reproducibility (5 days) D-IAP1 phynotypes: positive control Scatter plot of quantitative genomewide RNAi phenotypes Correlation=0.86 Efficiency of the screen: Negative control Positive control z = 2.9 +/- 0.8 • Averaged RNAi phenotypes of 72 genes encoding all annotated ribosomal proteins were tested. Further analysis of essential gene functions: Sequence-based Classification of Genes Quantitative Reproducibility • Phenotypes for a highly represented functional group of genes (nearly all of the tested genes encoding ribosomal components exhibited similar quantitative phenotypes). • Analysis of the phenotypes from independent dsRNAs targeting the same gene (show similar phenotypes). Results of Quantitative analysis • Identified many uncharacterized genes with essential cellular roles (e.g, HDC14318 with z=6.8 and 7.3). • Identified uncharacterized genes with phenotypes quantitatively similar to that of DIAP1 (z score 5). Examples: CG11700, a ubiquitin-like gene, and CG15455, a gene encoding an AML1-like transcription factor (z scores 7.2 and 7.4 in Kc167 cells). Classification of quantitative RNAi phenotypes of selected genes Flow cytometry analysis of propidium iodide stained DNA after 3 days RNAi Identification of anti-apoptotic gene function by severity of RNAi viability phenotypes Results • It suggests that the CG11700 gene may act in the same pathway as D-IAP1 to directly prevent Nc caspase-activated apoptotic cell death. • In contrast, a set of essential transcription factors may regulate complex responses for cell fate, proliferation, and/or cell survival that directly or indirectly initiate a partially caspasedependent apoptotic program. Critiques • Supplementary data !!!!!! Scatter plot of quantitative genome-wide RNAi phenotypes “For example, D-IAP1 resulted in a severe phenotype with z scores of 6.1 and 7.0 in S2R and Kc167 cells, respectively”. RNAi against ribosomal genes show similar quantitative phenotypes 34 genes: Similar quantitative RNAi phenotype putative falsenegative RNAi experiments Total number Of annotated as ribosomal subunits Lucimnescent cell viability assay • The assay determines the number of viable cells in culture by quantifying the amount of ATP present - a marker of metabolically active cells. The assay is designed for 96 or 384 well formats, making it ideal for automated high throughput screening (HTS), of cell proliferation and cytotoxicity assays. A homogeneous 'add, mix and measure' format makes the CellTiter-Glo assay simple to use with no cell washing, media removal or multiple pipetting steps. Measurements can be made on a CCD camera imaging device or a plate reading luminometer (no injector is required) as little as 10 minutes after addition of the reagent. • • • • The CellTiter-Blue™ Cell Viability Assay is a homogeneous, fluorometric method for estimating the number of viable cells present in a cell population. Similar to a video camera, digital camera use the lens which focuses the image onto a Charge Coupled Device (CCD), which then converts the image into electrical pulses. These pulses are then saved into memory. In short, Just as the film in a conventional camera records an image when light hits it, the CCD records the image electronically. • Z scores • Indicates how far and in what direction an item deviates from its distribution's mean. • Especially useful when seeking to compare the relative standings of items from distributions with different means and/or different standard deviations. • The number of standard deviations that a measurement is from the mean. This quantity indicates the relative location of a measurement within its distribution.