* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download MONOHYBRID CROSS
DNA sequencing wikipedia , lookup
Western blot wikipedia , lookup
DNA barcoding wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Maurice Wilkins wikipedia , lookup
Molecular evolution wikipedia , lookup
Restriction enzyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding DNA wikipedia , lookup
SNP genotyping wikipedia , lookup
Molecular cloning wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
DNA supercoil wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
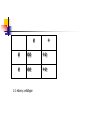
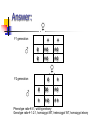
IDENTIFICATION OF POLYMORPHIC ALLELES 14.04.09 Quiz If you are to prepare a %3 agarose gel, what should be the amount of agarose in the 75 ml 1xTAE buffer? Show your calculations. If you cross a heterozygous wildtype with an ebony mutant what will be the phenotypic ratio of your F1 generation? Quiz If you are to prepare a %3 agarose gel, what should be the amount of agarose in the 75 ml 1xTAE buffer? Show your calculations. In 75 ml for %1 percent: 0,75 gr %3 percent: 2,25 gr e + e ee +e e ee +e 1:1 ebony: wildtype Answer: F1 generation e e F2 generation + +e +e + +e +e e + e ee +e + +e ++ Phenotype ratio3:1, wild-type:ebony Genotype ratio 1:2:1, homozygot WT, heterozygot WT, homozygot ebony Today’s experiments: 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. You will work as 4 groups First prepare 2% agarose gel Weigh 0,8 gr agarose into a flask Put 40ml 1X TBE buffer Dissolve the agarose completely by heating in the microwave When boils, remove the flask from the microwave Waits until it cools to 55C Add 2.5µl EtBr and mix Pour the melted agarose into gel apparatus Let it to harden Mix 5 µl of loading dye+ 5 µl of DNA sample, load on agarose gels Run agarose gels at 150 V for 15 min. Observe under UV light Electropherosis: 1. 2. A method to seperate, identify and purify DNA fragments 2 types of gels: Agarose gels Polyacrylamide gels Agarose Gels: From sea weed Cheap, non—toxic Low resolving power, but a higher range (200bp-50kb) Run in horizantal configuration In order to observe, EtBr is used EtBr intercalates DNA and it flouresces under UV light, so we can detect the location of the DNA fragments on the gel Agarose Gels: Agarose gel electrophoresis unit Agarose Gels: Polyacrylamide gels: Highly toxic, synthetic chemicals Prepared with acrylamide and bisacrylamide. In the presence of free radicals, it polymerizes into long chains Polyacrylamide gels: By changing acrylamide and bisacrylamide ratio, you can change the size of the pores Polyacrylamide gels: High resolution power, but a shorter range (5bp-500bp) Vertical configuration Polyacrylamide gels: Polyacrylamide gels: In order to visualize DNA, silver staining method can be used Ag+ ions bind to (-)ly charged DNA, Reduced to Ag which has a brown color Polymorphisms: 1. 2. Common variation in DNA sequence It is a kind of variation related to biodiversity, genetic variation, and adaptation Presence of more than one genetically distinct type in a single population Useful tools in genetic studies for linkage analysis, prenatal diagnosis, criminal cases and paternity tests RFLP (restriction fragment length polymorhism) VNTR (variable number of tandem repeats) RFLP: Restriction enzymes can recognize and cut specific DNA sequences Ex: Msp I enzyme can recognize CCGG Cfo I enzyme can recognize GCGC EcoR I can recognize GAATTC RFLP: -/- +/- +/+ RFLP: VNTR: (variable number of tandem repeats) Can be found on many chromosomes, and often show variations in length between individuals Each variant acts as an inherited allele, can be used for personal or parental identification Their analysis is useful in genetics and biology research, forensics, and DNA fingerprinting. Today’s experiments: 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. You will work as 4 groups First prepare 2% agarose gel Weigh 0,8 gr agarose into a flask Put 40ml 1X TBE buffer Dissolve the agarose completely by heating in the microwave When boils, remove the flask from the microwave Waits until it cools to 55C Add 2.5µl EtBr and mix Pour the melted agarose into gel apparatus Let it to harden Mix 5 µl of loading dye+ 5 µl of DNA sample, load on agarose gels Run agarose gels at 150 V for 15 min. Observe under UV light Today’s experiments: You will make a paternity test 1) On each bench, you have 5 DNA samples: mother, child and 3 father candidates 2) We will identify the father by checking 2 polymorhisms on different chromosomes 1. group RFLP (on chromosome 2) 2. group VNTR (on chromosome 5) Today’s experiments: 1. group RFLP (on chromosome 2) DNA fragment includes a Single nucleotide polymorhism (G or T) Msp I enzyme CCGG CCGG 600bp CCTG Digest with Msp I 400bp 200bp 600bp Today’s experiments: marker On agarose gel: load 3 µl 100bp marker mother child father3 father1 father2 600 500 bp 400 300 bp 200 bp 100 -/- +/- +/+ +/- -/- Today’s experiments: 2. group VNTR (on chromosome 5) DNA fragment includes different number of (CA) repeats You dont need to load marker Expected results: 800 700 600 RFLP: 1. group: mother child marker father3 father1 father2 616bp 500 400bp 400 300 200bp 200 100 -/- +/- -/- +/+ +/- VNTR: 2. group: mother child marker father3 father1 father2 800 4 700 600 3 500 400 2 300 200 1 100 2/4 1/2 2/6 1/2 1/3 Monohybrid cross results Ebony vs. wildtype Hypothesis: Body color is an autosomal trait and ebony type is recessive to the wild type” To test the accuracy of our hypothesis, we need to calculated to which extent our observed results are departed from the expected results Count Ebony vs. Wildtype Drosophila F1 generation e e F2 generation + +e +e + +e +e e + e ee +e + +e ++ Phenotype ratio3:1, wild-type:ebony Genotype ratio 1:2:1, homozygote WT, heterozygote WT, homozygote ebony