* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Fruit-specific RNAi-mediated suppression of DET1 enhances
Gene desert wikipedia , lookup
Gene nomenclature wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biochemical cascade wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA interference wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
RNA silencing wikipedia , lookup
Gene regulatory network wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene expression wikipedia , lookup
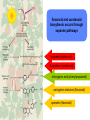
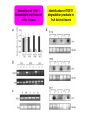
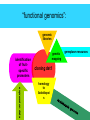
ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development The Plant Journal, Volume 39, Number 5, September 2004, pp. 697-714(18) www.pulsephotonics.com Rob Alba; Zhangjun Fei; Paxton Payton; Yang Liu; Shanna L. Moore; Paul Debbie; Jonathan Cohn; Mark D'Ascenzo; Jeffrey S. Gordon; Jocelyn K. C. Rose; Gregory Martin; Steven D. Tanksley; Mondher Bouzayen; Molly M. Jahn; Jim Giovannoni = transcribed portion of a genome expression profiling ONE GENE AT A TIME • RNA gel blot (=northern) • differential display NOT QUANTITATIVE, difficult to confirm • cDNA-AFLP CHEAP! little genomic data required SENSITIVE TO LOW-ABUNDANCE TRANSCRIPTS •NOT sequencing cDNA libraries (ESTs) EXPENSIVE AND LABOR-INTENSIVE • SAGE • microarrays expensive many genes at once, semi-quantitative gene discovery expressio n arrays “transcriptom e activity” mapping, coding regions EXPRESSED SEQUENCE TAGS lowabundance transcripts sequence errors 2 structure impairs RT human error the TOM1 array cDNA microarray based on EST library 12,899 features representing 8500 tomato genes protocols and confirmatory data available at The Tomato Expression Database: http://ted.bti.cornell.edu differential expression between tomato and pepper pericarp microarray pitfalls • cross-hybridization with related sequences • non-detection of sequence not included in the array • data handling is complex, therefore prone to human error (transformation, normalization, visualization, interpretation) • poor replication/experimental design • cDNA microarrays: – chimeric clones – inconsistent hybridization due to non-uniformity of microarray features artifacts occur 21 RUBISCO homologs but the pattern is robust photosynthesis-associated genes data visualization interpretation is problematic expression profiling simultaneously measures as much of the transcriptome as is represented on the chip this provides a valuable resource for studying regulatory and metabolic networks massive quantities of data are generated (and need to be analyzed) high costs and statistical difficulties encourage more focused approaches, but you only find what you’re looking for! Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes Ganga Rao Davuluri, Ageeth van Tuinen, Paul D Fraser, Alessandro Manfredonia, Robert Newman, Diane Burgess, David A Brummell, Stephen R King, Joe Palys, John Uhlig, Peter M Bramley, Henk M J Pennings & Chris Bowler Nature Biotechnology 23, 890 - 895 (2005) carotenoids • hydrophobic • mevalonic acid pathway flavonoids • hydrophilic • acetate-malonate pathway free radical scavengers/antioxidants enhance vertebrate immune system not synthesized by animals RANK IN NUTRIENT CONTRIBUTION TO AVERAGE U.S. DIET RANK IN NUTRIENT CONTENT modified from C.M. Rick flavonoid and carotenoid biosythesis occurs through separate pathways lycopene (carotenoid) B-carotene (carotenoid) chlorogenic acid (phenylpropanoid) naringenin-chalcone (flavonoid) quercetin (flavonoid) attempts at increasing phenolic/carotenoid production via expression of biosynthetic enzymes/transcription factors Niggeweg 2004: overexpression of HQA to increase CGA production in tomato Muir 2001: overexpression of petunia chalconeisomerase increases flavonol tomato Bovy 2002: increased flavonol production through heterologous expression of maize transcription factor Ye et al. 2000: production of β-carotene in rice endosperm via transformation with biosynthetic enzymes from daffodil, Erwinia Fraser et al. 2002: fruitspecific expression of Erwinia phytoene synthase increases carotenoid production in tomato Ducreux 2005: enhanced carotenoid production in potato via heterologous expression of Erwinia phytoene synthase Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. A C Mustilli, F Fenzi, R Ciliento, F Alfano, and C Bowler Plant Cell. 1999 February; 11(2): 145–157. de-etiolated 1: in A. thaliana, display light-grown phenotype when grown in the dark tomato hp-2 shows no phenotype in dark, but is hyper-responsive to light and has elevated pigment Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing Ganga Rao Davuluri, Ageeth van Tuinen , Diane Burgess, David A. Brummell, Stephen R. King, Joe Palys, John Uhlig, Henk M. J. Pennings, Chris Bowler, Anna Chiara Mustilli, Alessandro Manfredonia Robert Newman WT phenotypes consistent with loss of function, suggesting silencing HIGH PIGMENT! gene-specific methylation transgeneinduced silencing degradatio n of gene transcripts Davuluri et al. (2005) apply a post-tanscriptional silencing approach (RNAi) under a fruit-specific promoter dimunition of TDET1 transcript in fruit but not other tissues identification of TDET1 degradation products in fruit but not leaves lycopene B-carotene fruit weight brix “functional genomics”: genomic libraries identification of fruitspecific promoters genetic mapping cloning det1 expression data homology to Arabidopsi s germplasm resources