* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Angelman Syndrome (AS) and UBE3A (E6-AP)
Public health genomics wikipedia , lookup
Behavioral epigenetics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Human genetic variation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Oncogenomics wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Human genome wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Gene expression programming wikipedia , lookup
Human–animal hybrid wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of genetic engineering wikipedia , lookup
X-inactivation wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Gene expression profiling wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Designer baby wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Genome (book) wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics of human development wikipedia , lookup
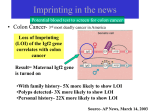
Epigenetics and Imprinting Dr Una Fairbrother Imprinting Describes the differential expression of genetic material at chromosomal/allelic level, depending on that material being maternal or paternal in origin. What is the imprint at a molecular level? An imprint is an heritable tag Usually associated with differential DNA methylation The met CpG usually on the silent chromosome In addition, histone aceylation and methylation has been identified Imprinting involves 3 distinct biological stages (a) Establishment of the imprint in gametes, according to the sex of the individual (b) Its maintenance during embryogenesis/adult somatic tissues (c) Erasure in the early germline Schematic of Imprinting Why imprinting? The advantages of biparental inheritance seem clear from the recessive nature of many disease mutations. Imprinting could help to maintain sexual reproduction. Imprinting in Development In mammals, imprinted genes have a vital role in development of the embryo and neonate. The division is not clear cut but; placental development appears to rely more heavily on paternal expression embryonic growth appears to rely more heavily on maternal expression. Imprinted Genes Present a complex Picture Of Expression In embryonic development, monoallelic expression is not always coincident with the onset of gene expression Can vary with development and differentiation. Imprinting, primary inactivation? Some evidence suggests that the imprint itself does not have to be a primary inactivation event The imprinting mechanism may involve additional trans-acting molecules. Imprinting and Disease Approx 80 imprinted genes in the human genome poss up to 600! Non-mendelian inheritance pattern with parent of origin effects Best characterised syndromes: Angelman/Prader-Willi syndromes on 15q (UBE3A- AS) Beckwith-Wiedemann syndrome (BWS) 11p (IGF2) Species Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Human Chr 01 01 06 06 06 07 07 07 07 07 07 07 11 11 11 11 11 11 11 11 11 11 11 11 11 11 11 Gene ARHI, NOEY2 TP73 PLAGL1; ZAC; LOT1 HYMAI IGF2R, M6PR (disputed) GRB10, MEG1 PEG10 DLX5 MEST, PEG1, MESTIT1, PEG1AS COPG2 (disputed PEG1-AS, MESTIT1 CPA4 WT1 (disputed) H19 IGF2 IGF2AS, PEG8 INS, insulin TRPM5, LTRPC5, MTR1 KCNQ1, KvLQT1 KCNQ1OT1, LIT1, KvLQT1-AS, KvDMR1 KCNQ1DN, BWRT CDKN1C, p57KIP2 SLC22A1L, IMPT1, BWR1A, ORCTL2, ITM, TSSC3, IPL, BWR1C, TSSC5, HET ,ZNF215 2G3-8 SDHD Human Human 14 14 15 Human Human Human 15 15 15 15 15 15 15 15 15 15 15 15 15 15 18 19 20 20 Human Human Human Human MEG3, GTL2 DLK1, PEG9 Paternally expressed noncoding transcripts on 15p (Prader-Willi syndrome region) MKRN3, ZNF127 NDN MAGEL2, NDNL1 SNRPN, SNURF PAR-SN HBII-13 PAR5, D15S226E HBII-85, PWCR1 IPW PAR1, D15S227E HBII-52 UBE3A, E6-AP, UBE3A-AS ATP10A, ATP10C Elongin A3 PEG3 NNAT, Neuronatin GNAS1, Gs alpha, NESP55, XLalpha s, GNAS-AS Features Of Imprinted Genes No direct sequence homology has been found among imprinted genes. Sequences carrying mat and pat gametic imprints resemble CpG islands containing GC rich regions of between 200 and 1500bp with a balanced CG:GC ratio. Imprinted sequences contain or are closely associated with a region of direct repeats ranging in size between 25 and 120bp. They are found in all imprinted genes analysed so far and are evolutionarily conserved. Imprinting Clusters Imprinted genes are clustered Best understood on chromosome 15 and 11 Complex coordinated regulation based on an imprinting centre (IC) or an imprinting control element (ICE) Coodinates in cis expression of neighbouring genes in one domain Action of Imprinting Centres Allele discriminating action of imprinting centres based on epigenetic modifications of chromatin Typically via methylation of cytosines (DNA)and histone acetlylation and methylation Acetylation of histones associated with transcriptionally active chromatin Methylation of histones and DNA associated with inactivation of transcription Imprinting and Transcription Methylation and acetylation lead to allele specific accessibility to transcriptional machinery And to DNA binding proteins acting as enhancers or insulators Setting and stability of imprinted gene expression controlled by ICs with multiple levels of DNA and chromatin modifications More complexity: IGF2 IGF2 (chromosome 11) has been shown to be imprinted very early on, in 8 cell human embryos recruits different promoters to confer mono or biallelic expression at different developmental stages. ….And KVLQT1 KVLQT1 (chromosome 11) has been reported as having a number of alternatively spliced transcripts shows both mono and biallelic expression patterns in different tissues Important in identification of imprinted genes associated with imprinted disorders Conservation Obviously evolutionary conservation is a good way to identify fundamentally important genes and sequence motifs A horse mare-donkey cross gives rise to a mule but a donkey-stallion gives rise to a hinny The callipyge gene phenotype “beautiful buttocks” is only expressed when paternally inherited Hum Mol Genet 1998 7:No10 review Imprinting in animal models Once thought to be restricted to mammals, genomic imprinting has been documented in angiosperm plants 1970, zebrafish 1995, insects, and C. elegans 2004 There may be genomic imprinting in drosophila, but transgenes have shown that imprinting switch regions act as silencers in flies In marsupials methylation on the X chr preferentially inactivates paternal X Mouse studies are one of the commonest in the literature Of Mice and Men Mouse models have been helpful in identifying the genes involved in Prader-Willi and Angelman syndrome The PWS mouse model has a partial maternal duplication of the region of mouse chromosome 7 homologous to human 15q11-13 The AS mouse model has a paternal duplication for the same region Imprinting in Mice Not Identical There are differences in imprinting between mice and humans Imprinting of H19, Igf2, p57kip2 and Snrpn is the same IGF2R appears to be monoallelic in mice and biallelic in humans, though this may be polymorphic Imprinting on Human Chromosome 15 IC regulates chromatin structure, DNA methylation and gene expression in a 2Mb region in 15q11-13 Mutations in IC cause chromosome to be stuck as a single gender and not assume the sex imprint of its ‘host’. >7 imprinted transcripts in AS region, paternally expressed UBE3A is only coding transcript maternally expressed Angelman Syndrome (AS) incidence of 1/20,000 live births severe mental retardation Lack of speech (commonly only a three to six word vocabulary) Ataxic gait Hand flapping Happy disposition including inappropriate laughter Diagnostic EEG Others Inheritance pattern of AS AS gene expressed MATERNALLY Chromosome 15 deletion in As and PWS Initially identified by the same cytogenetic lesion i.e. the deletion of 15q11-13 Prader-Willi Syndrome (PWS) Clinically distinct syndrome frequency of about 1 in 25,000 and probably the most common syndromal cause of human obesity Clinical features including: Mild mental retardation Obesity Short stature AS Oppositely Imprinted to PWS How Does AS Arise? AS caused by loss of function of a gene/genes from the maternal chromosome 15q11-13 (ICH study) 60% AS patients have large cytogenetic deletion 4% have uniparental disomy 4% have mutations in the imprinting centre (IC) 5% remaining, screened for mutations in the AS gene, UBE3A (E6AP) (50% familial, 10 sporadic) AS Oppositely Imprinted to PWS Recurrence risk typical large deletions are de novo and are expected to have less than 1% risk paternal UPD, (no parental translocation), less than 1% transmission of a structurally or functionally unbalanced chromosome complement can lead to 15q11-q13 deletions or to UPD and will result in case-specific recurrence risks. no large deletion or UPD, probably 50% due to maternal IC or UBE3A mutation see overhead* Risks confounded by mosaicism UBE3A (E6AP) and AS UBE3A (ubiquitin protein ligase) complex expression patterns. 5’ end of gene alternatively spliced in a variety of human tissue cell lines and in human foetal tissues including brain > 7 isoforms have been identified; monoallelic expression is tissue specific and isoform specific Human fibroblast and lymphoblast tissues and also adult mouse tissues show biallelic expression. Monoallelic expression of UBE3A Monoallelic expression of an isoform found in mouse brain tissue depression of expression is also evident in the hippocampus of a paternal UPD mouse Evidence for decrease of expression in human brain Clinical description of AS indicates a developmental brain disorder and so differential expression of the gene would be expected in brain tissue. UBE3A Has Even More Complex Splicing Normal UBE3A decreased in AS brain (MATERNAL) A new larger transcript decreased in PW brain (PATERNAL) Larger transcript is antisense and spans half UBE3A with additional down stream sequence Also another sense strand was detected which lies in between the 2nd and 3rd polyadenylation signal of UBE3A Imprinted and transcribed in the same way as UBE3A All other tissues antisense transcript not expressed The expression of the antisense transcript in brain may force the UBE3A transcript to be monoallelic in brain Some as yet uncharacterised AS patients could have a mutation in either the down stream sense transcript or the antisense transcript, see overhead* Mouse models Mouse models have been used to identify imprinted regions by engineering UPDs for mouse chromosomes, in part or in full Extensive mapping of imprinted mouse genes and their human homologues has been undertaken http://www.mgu.har.mrc.ac.uk/impri nting Similarities to Human Disease AS mouse: Neuro behavoural differences, mild ataxia, abnormal limb clasping, hyperactivity, diminished brain weight (10% smaller) The PWS mouse: increase in postnatal loss, small skeleton, grossly obese by 6 months Differences to Human disease AS mouse no detection of language difficulties (obviously), mouse also showed gross obesity - could be present in a subset of AS patients as late onset PWS mouse no detection of mild mental retardation. Usefulness of AS Mouse Model When gene was identified, AS mouse brains could be looked at and a monoallelically expressed transcript of UBE3A was found to be decreased as compared to the normal and the PWS mouse Outstanding questions How and when during germline development are old imprints removed and new ones introduced? Which demethylating activities and chromatin factors are involved? How does the spreading of epigenetic information in clusters work, - germline-specific, postzygotic phenomenon or both? How are imprints maintained when there is genome-wide active and passive demethylation in the early embryo? How many fundamentally different arrangements of imprinted genes and imprinting control elements are there in the genome? Cont… How conserved is imprinting between mammalian species? How precisely do imprinted genes affect extraembryonic and embryonic development, and the nutritional exchange with the mother? Are there interactions of imprinted genes (particularly antagonistic ones) in known, or in novel, physiological pathways? In addition to growth and behaviour, are there other developmental processes and mechanisms in which imprinted genes have a decisive role, and how will these fit with evolutionary theories?