* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Calvin Cycle
Biosynthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Cyanobacteria wikipedia , lookup
Magnesium in biology wikipedia , lookup
Chloroplast DNA wikipedia , lookup
Biochemistry wikipedia , lookup
Metalloprotein wikipedia , lookup
Electron transport chain wikipedia , lookup
Bioluminescence wikipedia , lookup
Chloroplast wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Microbial metabolism wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Citric acid cycle wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Light-dependent reactions wikipedia , lookup
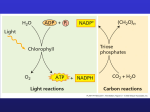
Advanced Biochemistry for Biotechnology, Photosynthesis: Calvin Cycle AN OVERVIEW OF PHOTOSYNTHESIS (Light reactions) • THE COLOR OF LIGHT SEEN IS THE COLOR NOT ABSORBED • Chloroplasts absorb light energy and convert it to chemical energy Light Reflected light Absorbed light Transmitted light Chloroplast AN OVERVIEW OF PHOTOSYNTHESIS • Photosynthesis is the process by which autotrophic organisms use light energy to make sugar and oxygen gas from carbon dioxide and water Carbon dioxide Water Glucose PHOTOSYNTHESIS Oxygen gas AN OVERVIEW OF PHOTOSYNTHESIS • The light reactions convert solar energy to chemical energy Light Chloroplast – Produce ATP & NADPH • The Calvin cycle makes sugar from carbon dioxide – ATP generated by the light reactions provides the energy for sugar synthesis – The NADPH produced by the light reactions provides the electrons for the reduction of carbon dioxide to glucose NADP ADP +P Light reactions Calvin cycle Chloroplasts: Sites of Photosynthesis • Photosynthesis – Occurs in chloroplasts, organelles in certain plants – All green plant parts have chloroplasts and carry out photosynthesis • The leaves have the most chloroplasts • The green color comes from chlorophyll in the chloroplasts • The pigments absorb light energy Photosynthesis occurs in chloroplasts • In most plants, photosynthesis occurs primarily in the leaves, in the chloroplasts • A chloroplast contains: – stroma, a fluid – grana, stacks of thylakoids • The thylakoids contain chlorophyll – Chlorophyll is the green pigment that captures light for photosynthesis • The location and structure of chloroplasts Chloroplast LEAF CROSS SECTION MESOPHYLL CELL LEAF Mesophyll CHLOROPLAST Intermembrane space Outer membrane Granum Grana Stroma Inner membrane Stroma Thylakoid Thylakoid compartment Chloroplast Pigments • Chloroplasts contain several pigments – Chlorophyll a – Chlorophyll b – Carotenoids Figure 7.7 Chlorophyll a & b •Chl a has a methyl group •Chl b has a carbonyl group Porphyrin ring delocalized e- Phytol tail Different pigments absorb light differently Excitation of chlorophyll in a chloroplast e 2 Less energy translates into longer wavelength. Excited state Heat Light Light (fluorescence) Photon Ground state Loss of energy due to heat causes the photons of light to be less energetic. Energy = (Planck’s constant) x (velocity of light)/(wavelength of light) Transition toward the red end of the visible spectrum. Chlorophyll molecule (a) Absorption of a photon (b) fluorescence of isolated chlorophyll in solution Cyclic Photophosphorylation • Process for ATP generation associated with some Photosynthetic Bacteria • Reaction Center => 700 nm • Two types of photosystems cooperate in the light reactions ATP mill Water-splitting photosystem NADPH-producing photosystem Noncyclic Photophosphorylation • Photosystem II regains electrons by splitting water, leaving O2 gas as a by-product Primary electron acceptor Primary electron acceptor Photons Energy for synthesis of PHOTOSYSTEM I PHOTOSYSTEM II by chemiosmosis Plants produce O2 gas by splitting H2O • The O2 liberated by photosynthesis is made from the oxygen in water (H+ and e-) How the Light Reactions Generate ATP and NADPH Primary electron acceptor Primary electron acceptor Energy to make NADP 3 2 Light Light Primary electron acceptor 1 Reactioncenter chlorophyll Water-splitting photosystem 2 H + 1 /2 NADPH-producing photosystem In the light reactions, electron transport chains generate ATP, NADPH, & O2 • Two connected photosystems collect photons of light and transfer the energy to chlorophyll electrons • The excited electrons are passed from the primary electron acceptor to electron transport chains – Their energy ends up in ATP and NADPH Chemiosmosis powers ATP synthesis in the light reactions • The electron transport chains are arranged with the photosystems in the thylakoid membranes and pump H+ through that membrane – The flow of H+ back through the membrane is harnessed by ATP synthase to make ATP – In the stroma, the H+ ions combine with NADP+ to form NADPH • The production of ATP by chemiosmosis in photosynthesis Thylakoid compartment (high H+) Light Light Thylakoid membrane Antenna molecules Stroma (low H+) ELECTRON TRANSPORT CHAIN PHOTOSYSTEM II PHOTOSYSTEM I ATP SYNTHASE • A Photosynthesis Road Map Chloroplast Light Stroma NADP Stack of thylakoids ADP +P Light reactions Calvin cycle Sugar used for Cellular respiration Cellulose Starch Other organic compounds Review: Photosynthesis uses light energy to make food molecules • A summary of the chemical processes of photosynthesis Chloroplast Light Photosystem II Electron transport chains Photosystem I CALVIN CYCLE Stroma Cellular respiration Cellulose Starch LIGHT REACTIONS CALVIN CYCLE Other organic compounds Photosynthesis takes place in chloroplasts. It includes light reactions and reactions that are not directly energized by light. 2 outer membranes grana disks (thylakoids) stroma compartment Chloroplast Light reactions: Energy of light is conserved as “high energy” phosphoanhydride bonds of ATP reducing power of NADPH. Proteins & pigments responsible for the light reactions are in thylakoid (grana disc) membranes. Light reaction pathways will be not be presented here. 22 Calvin Cycle, earlier designated the photosynthetic "dark reactions," is now called the carbon reactions pathway: 2 outer membranes grana disks (thylakoids) stroma compartment Chloroplast The free energy of cleavage of ~P bonds of ATP, and reducing power of NADPH, are used to fix and reduce CO2 to form carbohydrate. Enzymes & intermediates of the Calvin Cycle are located in the chloroplast stroma, a compartment somewhat 23 analogous to the mitochondrial matrix. H2C O OPO32- C -O O C H C OH H H C H2C OH OPO3 C H2C OH OPO32- 2- Ribulose-1,5-bisphosphate (RuBP) 3-Phosphoglycerate (3PG) Ribulose Bisphosphate Carboxylase (RuBP Carboxylase), catalyzes CO2 fixation: ribulose-1,5-bisphosphate + CO2 2 3-phosphoglycerate Because it can alternatively catalyze an oxygenase reaction, the enzyme is also called RuBP Carboxylase/Oxygenase (RuBisCO). It is the most abundant enzyme on earth.24 H 2C 1 O OPO32 C2 H C OH H C OH 3 4 H2C5 H 2C OPO32 ribulose-1,5bisphosphate O H+ H OPO32 C CO2 C OH C OH H 2C OPO32 enediolate intermediate H 2C HO C OPO32 CO2 O H C O C OH H2O H 2C OPO32 H C C H 2C O OH OPO32 b-keto 3-phosphoglycerate intermediate (2) RuBP Carboxylase - postulated mechanism: Extraction of H+ from C3 of ribulose-1,5-bisphosphate promotes formation of an enediolate intermediate. Nucleophilic attack on CO2 leads to formation of a b-keto acid intermediate, that reacts with water and cleaves to form 2 molecules of 3-phosphoglycerate. 25 HO H OPO3 HO C CO2 O H C OH OH H C OH OPO3 C CO2 C C H2C 2 H2 C H2C OPO3 2 Proposed b-keto acid intermediate H2 C 2 OPO32 2-Carboxyarabinitol-1,5bisphosphate (inhibitor) Transition state analogs of the postulated b-keto acid intermediate bind tightly to RuBP Carboxylase and inhibit its activity. Examples: 2-carboxyarabinitol-1,5-bisphosphate (CABP, above right) & carboxyarabinitol-1-phosphate (CA1P).26 RuBP Carboxylase in plants is a complex (L8S8) of: RuBisCO PDB 1RCX RuBisCO PDB 1RCX 8 large catalytic subunits (L, 477 residues, blue, cyan) 8 small subunits (S, 123 residues, shown in red). Some bacteria contain only the large subunit, with the smallest functional unit being a homodimer, L2. Roles of the small subunits have not been clearly defined. There is some evidence that interactions between large & 27 small subunits may regulate catalysis. ribulose-1,5Large subunits within bisphosphate RuBisCO are arranged as antiparallel dimers, with the N-terminal domain of one monomer adjacent to the Cterminal domain of the other. Each active site is at an interface between monomers within a dimer, explaining the minimal requirement for a dimeric structure. PDB 1RCX 2L & 2S subunits of RuBisCO The substrate binding site is at the mouth of an ab-barrel domain of the large subunit. Most active site residues are polar, including some 28 charged amino acids (e.g., Thr, Asn, Glu, Lys). O Enz-Lys + NH3 + HCO3 Enz-Lys H N + H2O + H+ C O Carbamate Formation with RuBP Carboxylase Activation "Active" RuBP Carboxylase has a carbamate that binds an essential Mg++ at the active site. The carbamate forms by reaction of HCO3 with the e-amino group of a lysine residue, in the presence of Mg++. HCO3 that reacts to form carbamate is distinct from CO2 that binds to RuBP Carboxylase as substrate. Mg++ bridges between oxygen atoms of the carbamate & substrate CO2. 29 Binding of either RuBP or a transition state analog to RuBP Carboxylase causes a conformational change to a "closed" conformation in which access of solvent water to the active site is blocked. RuBP Carboxylase (RuBisCO) can spontaneously deactivate by decarbamylation. In the absence of the carbamate group, RuBisCO tightly binds ribulose bisphosphate (RuBP) at the active site as a “dead end” complex, with the closed conformation, and is inactive in catalysis. In order for the carbamate to reform, the enzyme must undergo transition to the open conformation. 30 RuBP Carboxylase Activase is an ATP hydrolyzing (ATPase) enzyme that causes a conformational change in RuBP Carboxylase from a closed to an open state. This allows release of tightly bound RuBP or other sugar phosphate from the active site, and carbamate formation. Since photosynthetic light reactions produce ATP, the ATP dependence of RuBisCO activation provides a mechanism for light-dependent activation of the enzyme. The activase is a member of the AAA family of ATPases, many of which have chaperone-like roles. RuBP Carboxylase Activase is a large multimeric protein complex that may surround RuBisCO while inducing the conformational change to the open state. 31 O O Photorespiration: O2 can compete with CO2 for binding to RuBisCO, especially when [CO2] is low & [O2] is high. C O O H C H2C OH OPO 3 3-phosphoglycerate C 2 H2C OPO 3 2 phosphoglycolate When O2 reacts with ribulose-1,5-bisphosphate, the products are 3-phosphoglycerate plus the 2-C compound 2-phosphoglycolate. This reaction is the basis for the name RuBP Carboxylase/Oxygenase (RuBisCO). 32 O O C O O H Photorespiration: Diagram C H2C OH OPO 3 3-phosphoglycerate C 2 H2C OPO 3 2 phosphoglycolate The complex pathway that partly salvages C from 2-phosphoglycolate, via conversion to 3-phosphoglycerate, involves enzymes of chloroplasts, peroxisomes & mitochondria. This pathway recovers 3/4 of the C as 3-phosphoglycerate. The rest is released as CO2. Photorespiration is a wasteful process, substantially reducing efficiency of CO2 fixation, even at normal 33 ambient CO2. O2C C CH2 + HCO3 OPO32 O2C C CH2 CO2 + Pi O phosphoenolpyruvate oxaloacetate (PEP) PEP Carboxylase Most plants, designated C3, fix CO2 initially via RuBP Carboxylase, yielding the 3-C 3-phosphoglycerate. Plants designated C4 have one cell type in which phosphoenolpyruvate (PEP) is carboxylated via the enzyme PEP Carboxylase, to yield the 4-C oxaloacetate. Oxaloacetate is converted to other 4-C intermediates that are transported to cells active in photosynthesis, where CO2 is released by decarboxylation. 34 C4 plants maintain a high ratio of CO2/O2 within photosynthetic cells, thus minimizing photorespiration. Research has been aimed at increasing expression of and/or inserting genes for C4 pathway enzymes, such as PEP Carboxylase, in C3 plants. 35 O O Phosphoglycerate Kinase C H C H2 C ATP ADP OPO 3 3-phosphoglycerate O C H OH 2 Glyceraldehyde-3-phosphate Dehydrogenase C H2 C OPO 32 NADPH NADP+ OH OPO 32 1,3-bisphosphoglycerate CHO H Pi C H2 C OH OPO 32 glyceraldehyde3-phosphate Continuing with Calvin Cycle: The normal RuBP Carboxylase product, 3-phosphoglycerate is converted to glyceraldehyde-3-P. Phosphoglycerate Kinase catalyzes transfer of Pi from ATP to the carboxyl of 3-phosphoglycerate (RuBP Carboxylase product) to yield 1,3-bisphosphoglycerate. 36 O O Phosphoglycerate Kinase C H C H2 C ATP ADP OPO 3 3-phosphoglycerate O C H OH 2 Glyceraldehyde-3-phosphate Dehydrogenase C H2 C OPO 32 NADPH NADP+ OH OPO 32 1,3-bisphosphoglycerate CHO H Pi C H2 C OH OPO 32 glyceraldehyde3-phosphate Glyceraldehyde-3-P Dehydrogenase catalyzes reduction of the carboxyl of 1,3-bisphosphoglycerate to an aldehyde, with release of Pi, yielding glyceraldehyde-3-P. This is like the Glycolysis enzyme running backward, but the chloroplast Glyceraldehyde-3-P Dehydrogenase uses NADPH as e donor, while the cytosolic Glycolysis enzyme uses NAD+ as e acceptor. 37 Continuing with Calvin Cycle: A portion of the glyceraldehyde-3-P is converted back to ribulose-1,5-bisP, the substrate for RuBisCO, via reactions catalyzed by: Triose Phosphate Isomerase, Aldolase, Fructose Bisphosphatase, Sedoheptulose Bisphosphatase, Transketolase, Epimerase, Ribose Phosphate Isomerase, & Phosphoribulokinase. Many of these are similar to enzymes of Glycolysis, Gluconeogenesis or Pentose Phosphate Pathway, but are separate gene products found in the chloroplast stroma. (Enzymes of the other pathways listed are in the cytosol.) The process is similar to Pentose Phosphate Pathway run backwards. 38 Summary of Calvin cycle: 3 5-C ribulose-1,5-bisP (total of 15 C) are carboxylated (3 C added), cleaved, phosphorylated, reduced, & dephosphorylated, yielding 6 3-C glyceraldehyde-3-P (total of 18 C). Of these: 1 3-C glyceraldehyde-3-P exits as product. 5 3-C glyceraldehyde-3-P (15 C) are recycled back into 3 5-C ribulose-1,5-bisphosphate. C3 + C3 C6 C3 + C6 C4 + C5 C3 + C4 C7 C3 + C7 C5 + C5 Overall 5 C3 3 C5 39 TI Overall: glyceraldehyde-3-P dihydroxyacetone-P 5 C 3 3 C5 AL, FB Enzymes: fructose-6-P TI, Triosephosphate TK Isomerase xyulose-5-P + erythrose-4-P AL, Aldolase FB, Fructose-1,6AL, SB bisphosphatase sedoheptulose-7-P SB, SedoheptuloseTK Bisphosphatase TK, Transketolase xylulose-5-P + ribose-5-P EP, Epimerase EP IS IS, Isomerase (3) ribulose-5-P PK, PhosphoPK 40 (3) ribulose-1,5-bis-P ribulokinase CHO H Summary of Calvin Cycle O C carbon dioxide O C H2C OH OPO 32 glyceraldehyde3-phosphate 3 CO2 + 9 ATP + 6 NADPH glyceraldehyde-3-P + 9 ADP + 8 Pi + 6 NADP+ Glyceraldehyde-3-P may be converted to other CHO: • metabolites (e.g., fructose-6-P, glucose-1-P) • energy stores (e.g., sucrose, starch) • cell wall constituents (e.g., cellulose). Glyceraldehyde-3-P can also be utilized by plant cells as carbon source for synthesis of other compounds such as 41 fatty acids & amino acids. 2 outer membranes grana disks (thylakoids) stroma compartment Chloroplast There is evidence for multienzyme complexes of Calvin Cycle enzymes within the chloroplast stroma. Positioning of many Calvin Cycle enzymes close to the enzymes that produce their substrates or utilize their reaction products may increase efficiency of the pathway. 42 Regulation of Calvin Cycle Regulation prevents the Calvin Cycle from being active in the dark, when it might function in a futile cycle with Glycolysis & Pentose Phosphate Pathway, wasting ATP & NADPH. Light activates, or dark inhibits, the Calvin Cycle (previously called the “dark reaction”) in several ways. 43 H2O OH + H + h Regulation by Light. Chloroplast stroma (alkaline) (acid inside thylakoid disks) Light-activated e transfer is linked to pumping of H+ into thylakoid disks. pH in the stroma increases to about 8. Alkaline pH activates stromal Calvin Cycle enzymes RuBP Carboxylase, Fructose-1,6-Bisphosphatase & Sedoheptulose Bisphosphatase. The light-activated H+ shift is countered by Mg++ release from thylakoids to stroma. RuBP Carboxylase (in stroma) requires Mg++ binding to carbamate at the active site. 44 Some plants synthesize a transition-state inhibitor, carboxyarabinitol-1-phosphate (CA1P), in the dark. RuBP Carboxylase Activase facilitates release of CA1P from RuBP Carboxylase, when it is activated under conditions of light by thioredoxin. 45 Thioredoxin f PDB 1FAA disulfide Thioredoxin is a small protein with a disulfide that is reduced in chloroplasts via light-activated electron transfer. 46 ferredoxinOx S | S FerredoxinThioredoxin Reductase thioredoxin thioredoxin ferredoxinRed SH SH During illumination, the thioredoxin disulfide is reduced to a dithiol by ferredoxin, a constituent of the photosynthetic light reaction pathway, via an enzyme Ferredoxin-Thioredoxin Reductase. Reduced thioredoxin activates several Calvin Cycle enzymes, including Fructose-1,6-bisphosphatase, Sedoheptulose-1,7-bisphosphatase, and RuBP Carboxylase Activase, by reducing disulfides in those enzymes to thiols. 47