* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download TCGR: A Novel DNA/RNA Visualization Technique
Cre-Lox recombination wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Polyadenylation wikipedia , lookup
List of types of proteins wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Genetic code wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Community fingerprinting wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Biosynthesis wikipedia , lookup
Molecular evolution wikipedia , lookup
Protein structure prediction wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Messenger RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
RNA interference wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding DNA wikipedia , lookup
RNA silencing wikipedia , lookup
Non-coding RNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
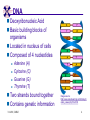
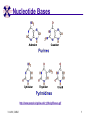
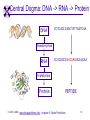
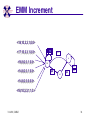
TCGR: A Novel DNA/RNA Visualization Technique Margaret H. Dunham and Donya Quick Southern Methodist University Dallas, Texas 75275 [email protected] Some slides presented at IEEE BIBE 2006 3/14/08, UMKC 1 Outline Introduction TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 2 Outline Introduction Background CGR/FCGR Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 3 DNA Deoxyribonucleic Acid Basic building blocks of organisms Located in nucleus of cells Composed of 4 nucleotides Adenine (A) Cytosine (C) Guanine (G) Thymine (T) Two strands bound together Contains genetic information 3/14/08, UMKC Image source: http://www.visionlearning.com/library/m odule_viewer.php?mid=63 4 Nucleotide Bases http://www.people.virginia.edu/~rjh9u/gif/bases.gif 3/14/08, UMKC 5 Transcription During transcription, DNA is converted in mRNA RNA is processed and noncoding regions removed Coding regions are converted in protein Enzyme (RNA Polymerase) that starts transcription by binding to DNA code 3/14/08, UMKC 6 Transcription http://ghs.gresham.k12.or.us/science/ps/sci/ ibbio/chem/nucleic/chpt15/transcripti on.gif 3/14/08, UMKC 7 RNA Ribonucleic Acid Contains A,C,G but U (Uracil) instead of T Single Stranded May fold back on itself Needed to create proteins Move around cells – can act like a messenger mRNA – moves out of nucleus to other parts of cell 3/14/08, UMKC 8 Translation Synthesis of Proteins from mRNA Nucleotide sequence of mRNA converted in amino acid sequence of protein Four nucleotides Twenty amino acids Codon – Group of 3 nucleotides Amino acids have many codings 3/14/08, UMKC 9 Central Dogma: DNA -> RNA -> Protein DNA CCTGAGCCAACTATTGATGAA transcription RNA CCUGAGCCAACUAUUGAUGAA translation Protein PEPTIDE 3/14/08, UMKC www.bioalgorithms.info; chapter 6; Gene Prediction 10 http://www.time.com/time/magazine/article/0,91 71,1541283,00.html 3/14/08, UMKC 11 Human Genome Scientists originally thought there would be about 100,000 genes Appear to be about 20,000 WHY? Almost identical to that of Chimps. What makes the difference? Answers appear to lie in the noncoding regions of the DNA (formerly thought to be junk) 3/14/08, UMKC 12 More Questions If each cell in an organism contains the same DNA – How does each cell behave differently? Why do cells behave differently during childhood development? What causes some cells to act differently – such as during disease? DNA contains many genes, but only a few are being transcribed – why? One answer - miRNA 3/14/08, UMKC 13 miRNA Short (20-25nt) sequence of noncoding RNA Single strand Previously assumed to be garbage Impact/Prevent translation of mRNA Bind to target areas in mRNA – Problem is that this binding is not perfect (particularly in animals) mRNA may have multiple (nonoverlapping) binding sites for one miRNA 3/14/08, UMKC 14 miRNA Functions Causes some cancers Embryo Development Cell Differentiation Cell Death Prevents the production of a protein that causes lung cancer Control brain development in zebra fish Associated with HIV 3/14/08, UMKC 15 Outline Introduction Background CGR/FCGR Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 16 Chaos Game Representation (CGR) Scatter plot showing occurrence of patterns of nucleotides. 3/14/08, UMKC University of the Basque Country http://insilico.ehu.es/genomics/my_words/ 17 FCGR Chaos Game Representation 2D technique to visually (CGR) see the distribution of subpatterns Our technique is based on the following: Generate totals for each subpattern Scale totals to a [0,1] range. (Note scaling can be a problem) Convert range to red/blue A CA C G UG U A CA C G UG U A CA C G UG U A CA C G UG U A CA C G UG U A CA C G UG U A CA C G UG U A CA C G UG U • 0-0.5: White to Blue • 0.5-1: Blue to Red 3/14/08, UMKC 19 FCGR Example Homo sapiens – all mature miRNA Patterns of length 3 UUC GUG 3/14/08, UMKC 21 Outline Introduction Background CGR/FCGR Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 22 Motivation 2000bp Flanking Upstream Region mir-258.2 in C elegans a) All 2000 bp 3/14/08, UMKC b) First 240 bp b) Last 240 bp 23 Research Objectives Identify, develop, and implement algorithms which can be used for identifying potential miRNA functions. Create an online tool which can be used by other researchers to apply our algorithms to new data. 3/14/08, UMKC 24 Outline Introduction CGR/FCGR miRNA Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 25 Temporal CGR (TCGR) Temporal version of Frequency CGR In our context temporal means the starting location of a window 2D Array Each Row represents counts for a particular window in sequence • First row – first window • Last row – last window • We start successive windows at the next character location Each Column represents the counts for the associated pattern in that window • Initially we have assumed order of patterns is alphabetic Size of TCGR depends on sequence length and subpattern lengt As sequence lengths vary, we only examine complete windows We only count patterns completely contained in each window. 3/14/08, UMKC 26 TCGR Example acgtgcacgtaactgattccggaaccaaatgtgcccacgtcga Moving Window Pos 0-8 Pos 1-9 A 2 1 C 3 3 G 3 3 T 1 2 4 2 1 C 0.6 0.6 G 0.6 0.6 T 0.2 0.4 0.8 0.4 0.2 … Pos 34-42 2 Pos 0-8 Pos 1-9 A 0.4 0.2 … Pos 34-42 0.4 3/14/08, UMKC 27 TCGR Example (cont’d) TCGRs for Sub-patterns of length 1, 2, and 3 3/14/08, UMKC 28 TCGR Example (cont’d) ACGT Window 0: Pos 0-8 Window 1: Pos 1-9 acgtgcacg cgtgcacgt Window 17: Pos 17-25 Window 18: Pos 18-26 tccggaacc ccggaacca Window 34: Pos 34-42 ccacgtcga 3/14/08, UMKC 29 TCGR – Mature miRNA (Window=5; Pattern=3) C. elegans Homo sapiens Mus musculus All Mature 3/14/08, UMKC ACG CGC GCG UCG 31 Outline Introduction CGR/FCGR miRNA Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 32 EMM Overview Time Varying Discrete First Order Markov Model Nodes are clusters of real world states. Learning continues during prediction phase. Learning: Transition probabilities between nodes Node labels (centroid of cluster) Nodes are added and removed as data arrives 3/14/08, UMKC 33 EMM Definition Extensible Markov Model (EMM): at any time t, EMM consists of an MC with designated current node, Nn, and algorithms to modify it, where algorithms include: EMMCluster, which defines a technique for matching between input data at time t + 1 and existing states in the MC at time t. EMMIncrement algorithm, which updates MC at time t + 1 given the MC at time t and clustering measure result at time t + 1. EMMDecrement algorithm, which removes nodes from the EMM when needed. 3/14/08, UMKC 34 EMM Cluster Find closest node to incoming event. If none “close” create new node Labeling of cluster is centroid of members in cluster O(n) 3/14/08, UMKC 35 EMM Increment <18,10,3,3,1,0,0> <17,10,2,3,1,0,0> <16,9,2,3,1,0,0> <14,8,2,3,1,0,0> 2/3 2/3 2/21 2/3 1/1 1/2 1/2 N3 N1 1/3 N2 1/1 1/2 1/1 <14,8,2,3,0,0,0> <18,10,3,3,1,1,0.> 3/14/08, UMKC 36 Outline Introduction CGR/FCGR miRNA Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 37 Research Approach 1. Represent potential miRNA sequence with TCGR sequence of count vectors 2. Create EMM using count vectors for known miRNA (miRNA stem loops, miRNA targets) 3. Predict unknown sequence to be miRNA (miRNA stem loop, miRNA target) based on normalized product of transition probabilities along clustering path in EMM 3/14/08, UMKC 38 Related Work 1 Predicted occurrence of pre-miRNA segments form a set of hairpin sequences No assumptions about biological function or conservation across species. Used SVMs to differentiate the structure of hiarpin segments that contained premiRNAs from those that did not. Sensitivey of 93.3% Specificity of 88.1% 1 C. Xue, F. Li, T. He, G. Liu, Y. Li, nad X. Zhang, “Classification of Real and Pseudo MicroRNA Precursors using Local Structure-Sequence Features and Support Vector Machine,” BMC Bioinformatics, vol 6, no 310. 3/14/08, UMKC 39 Preliminary Test Data1 Positive Training: This dataset consists of 163 human pre-miRNAs with lengths of 62-119. Negative Training: This dataset was obtained from protein coding regions of human RefSeq genes. As these are from coding regions it is likely that there are no true pre-miRNAs in this data. This dataset contains 168 sequences with lengths between 63 and 110 characters. Positive Test: This dataset contains 30 pre-miRNAs. Negative Test: This dataset contains 1000 randomly chosen sequences from coding regions. 1 C. Xue, F. Li, T. He, G. Liu, Y. Li, nad X. Zhang, “Classification of Real and Pseudo MicroRNA Precursors using Local StructureSequence Features and Support Vector Machine,” BMC Bioinformatics, vol 6, no 310. 3/14/08, UMKC 40 TCGRs for Xue Training Data P O S I T I V E N E G A T I V E 3/14/08, UMKC 41 TCGRs for Xue Test Data P O S I T I V E N E G AT I V E 3/14/08, UMKC 42 Predictive Probabilities with Xue’s Data EMM Mean Std Dev Max Min Negative Test-Neg 0 0 0 0 Test-Pos 0 0 0 0 Train-Neg 0.37963 0.050085 0.91256 0.2945 Train-Pos 0 0 0 0 Test-Neg 0 0 0 0 Test-Pos 0.25894 0.18701 0.42075 0 Train-Neg 0 0 Train-Pos 0.38926 0.048439 0.91155 0.32209 Positive 3/14/08, UMKC Test Data 0 0 43 Preliminary Test Results Positive EMM Cutoff Probability = 0.3 False Positive Rate = 0% True Positive Rate = 66% Test results could be improved by meta classifiers combining multiple positive and negative classifiers together. 3/14/08, UMKC 44 Outline Introduction CGR/FCGR miRNA Motivation Research Objective TCGR EMM miRNA Prediction using TCGR/EMM Conclusion / Future Work 3/14/08, UMKC 45 Future Research 1. Obtain all known mature miRNA sequences for a species – initially the 119 C. elegans miRNAs. 2. Create TCGR count vectors for each sequence and each sub-pattern length (1,2,3,4,5). 3. Train EMMs using this data for each sub-pattern length. Thus five EMMs will be created 4. Obtain negative data (much as Xue did in his research) from coding regions for C. elegans. 5. Train EMMs using this data for each sub-pattern length. Thus five EMMs will be created 6. Construct a meta-classifier based on the combined results of prediction from each of these ten EMMs. 7. Apply the EMM classifier to the existing ~75x106 base pairs of non-exonic sequence in the C. elegans genome to search for miRNAs. Note: all 119 validated C. elegans miRNAs are contained in the non-exonic part of the genome and thus the first pass of the algorithm will be tested for its ability to detect all 119 validated miRNAs. 8. Validate the prediction of novel miRNAs using molecular biology. 3/14/08, UMKC 46