* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
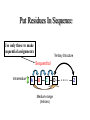
Download Use only these to make sequential assignments
List of types of proteins wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Circular dichroism wikipedia , lookup
Protein domain wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein folding wikipedia , lookup
Proteolysis wikipedia , lookup
Homology modeling wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
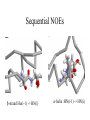
Resonance Assignment •NMR analysis of proteins •Sequential resonance assignment strategies •Practical Issues Sequential Assignment 1.Assign the protein backbone N, HN, CA, HA, CB, HB, C 2. Assign sidechains 3. Assign NOE contacts Assignment Strategy 1. Identify resonances for each amino acid 2. Put amino acids in order - Sequential assignment (R-G-S,T-L-G-S) 3. Place fragments in aa sequence - Sequence-specific assignment Put Residues In Sequence Use only these to make sequential assignments Tertiary Structure Sequential Intraresidue A B C Medium-range (helices) D •••• Z Sequential NOEs b-strand Ha(i-1) -> HN(i) a-helix :HN(i-1) -> HN(i) NOE based Assignment 15N edited 1H-1H TOCSY/NOESY Large Scalar Couplings Less Sensitive to MW of the Protein Superior to 1H homonuclear strategy: H-H couplings <20 Hz Mixing is faster so less time for signal to relax Heteronuclear Assignments: Backbone Experiments Names of scalar experiments based on atoms detected Acquired in ‘pairs’ Heteronuclear Assignments: Side Chain Experiments Determining Protein Fold In Structure Calculations 1. Determine secondary structure •CSI directly from assignments •Medium-range NOEs 2. Add key long-range NOEs to fold Identify more NOEs In the course of refinement