* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Pyruvate Dehydrogenase
Basal metabolic rate wikipedia , lookup
Adenosine triphosphate wikipedia , lookup
Lipid signaling wikipedia , lookup
Proteolysis wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Western blot wikipedia , lookup
Photosynthetic reaction centre wikipedia , lookup
Butyric acid wikipedia , lookup
Microbial metabolism wikipedia , lookup
Metalloprotein wikipedia , lookup
Mitochondrion wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Electron transport chain wikipedia , lookup
Biosynthesis wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Biochemistry wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Lactate dehydrogenase wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
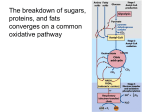
Molecular Biochemistry I Pyruvate Dehydrogenase Copyright © 1999-2006 by Joyce J. Diwan. All rights reserved. Glycolysis occurs in the cytosol of cells. Pyruvate enters the mitochondrion to be metabolized further. Mitochondrial Compartments: matrix cristae intermembrane space inner membrane mitochondrion outer membrane The matrix contains Pyruvate Dehydrogenase, enzymes of Krebs Cycle, and other pathways, e.g., fatty acid oxidation & amino acid metabolism. The outer membrane contains large VDAC channels, similar to bacterial porin channels, making the outer membrane leaky to ions & small molecules. matrix The inner membrane is the major permeability barrier of the mitochondrion. cristae intermembrane space inner membrane mitochondrion outer membrane It contains various transport catalysts, including a carrier protein that allows pyruvate to enter the matrix. It is highly convoluted, with infoldings called cristae. Embedded in the inner membrane are constituents of the respiratory chain and ATP Synthase. Pyruvate Dehydrogenase H3C O O C C pyruvate HSCoA O O H3C NAD+ NADH C S CoA + CO2 acetyl-CoA Pyruvate Dehydrogenase, catalyzes oxidative decarboxylation of pyruvate, to form acetyl-CoA. Pyruvate Dehydrogenase: a large complex containing many copies of each of 3 enzymes, E1, E2, & E3. The inner core of mammalian Pyruvate Dehydrogenase is an icosahedral structure consisting of 60 copies of E2. At the periphery of the complex are: • 30 copies of E1 (itself a tetramer with subunits a2b2). • 12 copies of E3 (a homodimer), plus 12 copies of an E3 binding protein that links E3 to E2. Pyruvate Dehydrogenase Subunits Enzyme Abbreviated Prosthetic Group Pyruvate Dehydrogenase E1 Thiamine pyrophosphate (TPP) Dihydrolipoyl Transacetylase E2 Lipoamide Dihydrolipoyl Dehydrogenase E3 FAD dimethylisoalloxazine O H C C N O H3C C C C NH H3C C C C C C H N H C + 2e +2H O N H N H3C C C C NH H3C C C C C C H CH2 FAD N O N H CH2 HC OH HC OH HC OH O H2C C O P O- Adenine O O P O Ribose FADH2 HC OH HC OH HC OH O H2C O O- P Adenine O O O- P O Ribose O- FAD (Flavin Adenine Dinucleotide is derived from the vitamin riboflavin. The dimethylisoalloxazine ring system undergoes oxidation/reduction. FAD is a prosthetic group, permanently part of E3. Reaction: FAD + 2 e- + 2 H+ FADH2 O O H3C CH2 N H3C CH2 H O P O O + N C N CH2 P O S acidic H+ NH2 thiamine pyrophosphate (TPP) Thiamine pyrophosphate (TPP) is a derivative of thiamine (vitamin B1). Nutritional deficiency of thiamine leads to the disease beriberi. Beriberi affects especially the brain, because TPP is required for CHO metabolism, & the brain depends on glucose metabolism for energy. O O O H3C CH2 N H3C CH2 H O P O O + N C N CH2 O P O S acidic H+ NH2 thiamine pyrophosphate (TPP) Thiamine pyrophosphate (TPP) mechanism: O O H+ readily dissociates from the C between N and S in the thiazole ring. C The resulting carbanion (ylid) can attack the electron-deficient keto carbon of pyruvate. CH3 C O pyruvate S CH2 CH2 S lipoic acid CH O CH2 CH2 CH2 CH2 C Lipoamide includes a dithiol that undergoes oxidation/ reduction. lipoamide lysine NH NH (CH2)4 CH C O 2e + 2H+ HS CH2 CH2 HS O CH CH2 CH2 CH2 CH2 C dihydrolipoamide NH NH (CH2)4 CH C O S CH2 CH2 S CH lipoic acid O CH2 CH2 CH2 CH2 C lipoamide lysine NH NH (CH2)4 CH C O 2e + 2H+ The carboxyl at the end of lipoic acid's hydrocarbon chain CH2 forms an HS amide bond to the side-chain amino group of a lysine residue ofCHE2 2, yielding lipoamide. NH HS CH O A long flexible arm, including hydrocarbon chains of CH2 CH 2 CH2 CH2 C NH (CH2)4 CH lipoate and the lysine R-group, links each lipoamide dithiol C O group to one of 2 lipoate-binding domains of each E2. Lipoate-binding domains are themselves part of a flexible strand of E2 that extends out from the core of the complex. S CH2 CH2 S CH lipoic acid O CH2 CH2 CH2 CH2 C lipoamide lysine NH NH (CH2)4 CH C O 2e + 2H+ The long flexible attachment allows lipoamide functional CH2 between E2 active sites in the core of the groups toHS swing CHsites 2 complex & active of E1 & E3 in the outer shell. NH HS CH O E3 binding protein that binds E3 to E2 also has attached CH2 CH2 CH2 CH2 C NH (CH2)4 CH lipoamide that can exchange of reducing equivalents with C O lipoamide on E2. Diagrams in: website of the laboratory of Wim Hol article by Milne et al. (Fig. 5) H2O HS R' As O S R' + HS As S R R Organic arsenicals are potent inhibitors of lipoamidecontaining enzymes such as Pyruvate Dehydrogenase. These highly toxic compounds react with “vicinal” dithiols such as the functional group of lipoamide. O C CH3 Coenzyme A-SH + HO acetic acid O Coenzyme A-S C CH3 + H2O acetyl-CoA In the overall reaction catalyzed by the Pyruvate Dehydrogenase complex, the acetic acid generated is transferred to coenzyme A. H O H H C The final electron acceptor is NAD+. C NH2 + N O NH2 + 2e + H N R R NAD+ NADH Sequence of reactions catalyzed by Pyruvate Dehydrogenase complex: 1. The keto C of pyruvate reacts with the carbanion of TPP on E1 to yield an addition compound. The electron-pulling (+) charged N of the thiazole ring promotes CO2 loss. Hydroxyethyl-TPP remains. 2. The hydroxyethyl carbanion on TPP of E1 reacts with the disulfide of lipoamide on E2. What was the keto C of pyruvate is oxidized to a carboxylic acid, as the lipoamide disulfide is reduced to a dithiol. The acetate formed by oxidation of the hydroxyethyl is linked to one of the thiols of the reduced lipoamide as a thioester (~). Sequence of reactions (continued) 3. Acetate is transferred from the thiol of lipoamide to the thiol of coenzyme A, yielding acetyl CoA. 4. The reduced lipoamide, swings over to the E3 active site. Dihydrolipoamide is reoxidized to the disulfide, as 2 e + 2 H+ are transferred to a disulfide on E3 (disulfide interchange). 3. The dithiol on E3 is reoxidized as 2 e- + 2 H+ are transferred to FAD. The resulting FADH2 is reoxidized by electron transfer to NAD+, to yield NADH + H+. View an animation of the Pyruvate Dehydrogenase reaction sequence. O H3C C S CoA acetyl-coenzyme A Acetyl CoA, a product of the Pyruvate Dehydrogenase reaction, is a central compound in metabolism. The "high energy" thioester linkage makes it an excellent donor of the acetate moiety. glucose-6-P Glycolysis pyruvate fatty acids acetyl CoA oxaloacetate ketone bodies cholesterol citrate Krebs Cycle Acetyl CoA functions as: input to Krebs Cycle, where the acetate moiety is further degraded to CO2. donor of acetate for synthesis of fatty acids, ketone bodies, & cholesterol. Regulation of Pyruvate Dehydrogenase Complex: Product inhibition by NADH & acetyl CoA: NADH competes with NAD+ for binding to E3. Acetyl CoA competes with CoA for binding to E2. Regulation by E1 phosphorylation/dephosphorylation: Specific regulatory Kinases & Phosphatases associated with Pyruvate Dehydrogenase in the mitochondrial matrix: Pyruvate Dehydrogenase Kinases catalyze phosphorylation of serine residues of E1, inhibiting the complex. Pyruvate Dehydrogenase Phosphatases reverse this inhibition. Pyruvate Dehydrogenase Kinases are activated by NADH & acetyl-CoA, providing another way the 2 major products of Pyruvate Dehydrogenase reaction inhibit the complex. Kinase activation involves interaction with E2 subunits to sense changes in oxidation state & acetylation of lipoamide caused by NADH & acetyl-CoA. During starvation: Pyruvate Dehydrogenase Kinase increases in amount in most tissues, including skeletal muscle, via increased gene transcription. Under the same conditions, the amount of Pyruvate Dehydrogenase Phosphatase decreases. The resulting inhibition of Pyruvate Dehydrogenase prevents muscle and other tissues from catabolizing glucose & gluconeogenesis precursors. Metabolism shifts toward fat utilization. Muscle protein breakdown to supply gluconeogenesis precursors is minimized. Available glucose is spared for use by the brain. A Ca++-sensitive isoform of the phosphatase that removes Pi from E1 is expressed in muscle. Pyruvate Dehydrogenase in matrix mitochondrion Ca++ The increased cytosolic Ca++ that occurs during activation of muscle contraction can lead to Ca++ uptake by mitochondria. The higher Ca++ stimulates the phosphatase, & dephosphorylation activates Pyruvate Dehydrogenase. Thus mitochondrial metabolism may be stimulated during exercise. Lecture notes relating to Krebs Cycle are not provided, because students will present the lectures. However see the summary of Krebs Cycle embedded in the class web page. Some questions on Krebs Cycle are included in the self-study quiz for this class.