* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download lecture 15
Immunoprecipitation wikipedia , lookup
Rosetta@home wikipedia , lookup
Structural alignment wikipedia , lookup
Protein design wikipedia , lookup
Circular dichroism wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Cooperative binding wikipedia , lookup
P-type ATPase wikipedia , lookup
Protein folding wikipedia , lookup
ATP-binding cassette transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
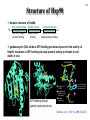
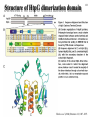
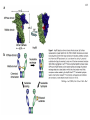
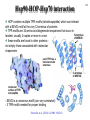
Hsp90 as a regulator of protein conformation and function Hsp90 is an abundant eukaryotic protein makes up about ~2% of cytosolic protein content not surprising: number of proteins it interacts with is huge ~90 kDa in size; forms dimers phosphoprotein Hsp90 is also present in the ER, where it is termed Grp94, or Glucoseregulated protein 94kDa (most abundant protein of the ER) production increased by cellular stresses, i.e. it is a heat-shock protein it is essential for viability in both cytosol and endoplasmic reticulum, where it is ubiquitously present Hsp90 exists in some bacteria but not archaea 15-1 15-2 Structure of Hsp90 domain structure of Hsp90: N-terminal domain middle domain N GA, ATP, target protein binding target protein binding C-terminal domain C dimerization site; target protein binding geldanamycin (GA) binds in ATP-binding pocket and prevents the activity of Hsp90; mutations in ATP binding site also prevent activity and leads to cell death in vivo ATP-binding domain (partial crystal structure) Stebbins et al. (1997) Cell 89, 239-250. Structure of HtpG dimerization domain 15-3 Harris et al. (2004) Structure 12, 1087-1097. Structure of HtpG 15-4 A Simplified Schematic of Possible Hsp90 Function Substrate is represented as a green circle to mark the expected position of contacts, but not as a direct indication of substrate size or structural details. The bulk of the substrate may, in fact, be variously extended outside of the Hsp90 clamp with chaperone:substrate contacts limited to a subdomain or smaller structural element of the client protein. Also, binding of ATP is known to stimulate the association of the amino-terminal domains, but as the timing of hydrolysis and release is not yet well understood, these details have been excluded from these figures for simplicity. Substrate is presumed to bind within the Hsp90 dimer clamp, contacting multiple mobile hydrophobic elements including helix 2 of the carboxy-terminal domains (shown as cylinders) and loops or patches along the inner surface of the middle domain (not explicitly shown). The association of the amino-terminal domains, stimulated by ATP binding, occludes the inner volume and juxtaposes the hydrophobic features. We show three possible routes of substrate release (blue arrows). Reversal of the initial binding event may return the chaperone to its open state with amino terminal domains separated (left). Alternately, full closure of the clamp may be incompatible with substrate binding as the hydrophobic features are mutually masked (upper right). Finally, inspired by the GHKL family member topoisomerases, transient dissociation of the Structure of Full-length Hsp90 (yeast Hsp90) carboxy-terminal domains could cause the exposed hydrophobic dimer interface to compete for binding with helix 2, thereby synchronizing substrate from Pearl and Promodou, Ann. Rev. Biochem. 2006 release with an opening of the Hsp90 topological ring to permit substrate to exit the Hsp90 clamp (lower right). 15-5 Stirling et al. (2006) Nat. Struct. Mol. Biol. Physiological targets of Hsp90 15-6 Hsp90 interacts with, regulates the conformation of, and the activity of, a large variety of cell signalling molecules, transcription factors, cytoskeleton, etc. Substrates heat-shock factor (HSF) Hsp90 downregulates activity in conjunction with Hsp70 system other transcription factors receptors (steroid, glucocorticoid), hypoxia-inducible factor-1 (HIF-1), etc. kinases tyrosine kinases (v-src, lck, insulin receptor, etc.) serine-thronine kinases (eIF-2 kinase, v-raf, c-raf, etc.) protein kinase CK-II cytoskeleton actin, tubulin (protection during heat stress) 15-7 Hsp90-associated proteins it is believed that Hsp90 never functions in isolation in eukaryotes; it always appears to be associated with a variety of cofactors Cofactor Notes Hsp70 Hsp90 activity dependent on Hsp70 system (incl. Hsp40) HOP HOP, Heat-shock Organizing Protein, brings Hsp70 and Hsp90 together via TPR interaction domains p23 modulates ATPase activity of Hsp90 HIP co-chaperone of Hsp70 PPIases cyclophilin-40, FKBP51, FKBP52 others kinase-targeting co-chaperone Cdc37/p50 15-8 Hsp90 functional cycle Hsp90 not only assists the folding of proteins, but can also modulate the conformation/function of proteins binding of steroid to Hsp90-bound steroid receptor releases the receptor in a form that can bind DNA and activate transcription Hsp40 target protein (non-native/ non-functional) HIP Hsp70 Hsp90 is also involved in quality control: binding of denatured protein can lead either to its folding or its degradation HIP Hsp90 p23 FKBP HIP Hsp70 Hsp70 HOP HOP p23 target protein (native/functional) p23 FKBP Hsp90 FKBP Hsp90-HOP-Hsp70 interaction HOP contains multiple TPR motifs (tetratricopeptide) which can interact with a EEVD motif at the very C-terminus of proteins TPR motifs are 34 amino acid degenerate sequences that occur in N-terminus tandem, usually 3 copies or more in a row of MEEVD these motifs are found in other proteins-not simply those associated with molecular chaperones each TPR has a helix-turn-helix structure C-terminus of MEEVD molecular surface of TPR and peptide - EEVD is a consensus motif (can vary somewhat) - 3 TPR motifs needed for proper binding Scheufler et al. (2000) Cell 101, 199-210. 15-9 TPR motif specificity of HOP Domain structure of HOP Example ITC data; incubate one component with another at different ratios, measure heat ITC data shows Hsp70/Hsp90 preference for TPR1 or TPR2A binding sites n (ratio of binding) is ~1 for all Kd measured (lower μM means tighter binding) 15-10 15-11 Is Hsp90 a ‘typical’ chaperone? Hsp90 can prevent the aggregation of a denatured protein upon dilution from a chaotrope (urea, guanidine hydrochloride) but it is not very efficient compared to other chaperones cannot refold a protein by itself how can it recognize so many different proteins that belong to rather limited classes (e.g. all sorts of different transcription factors, kinases, etc.) Susan Lindquist laboratory: Hsp90 as a capacitor for evolution