* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Amino acid metabolism III. Brake down of amino acids
Mitogen-activated protein kinase wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Butyric acid wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Catalytic triad wikipedia , lookup
Point mutation wikipedia , lookup
Glyceroneogenesis wikipedia , lookup
Proteolysis wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Peptide synthesis wikipedia , lookup
Protein structure prediction wikipedia , lookup
Metalloprotein wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Genetic code wikipedia , lookup
Biochemistry wikipedia , lookup
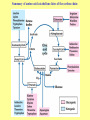
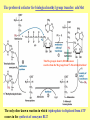
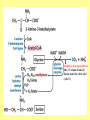
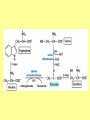
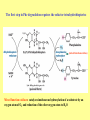
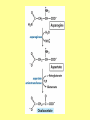
Amino acid metabolism III. Brake down of amino acids, glucoplastic and ketoplastic amino acids Figures: Lehninger-4ed; chapter: 18 Stryer-5ed; chapter: 23 Overview of amino acid catabolism in mammals Summary of amino acid catabolism: fates of the carbon chain Purely ketogenic amino acids: can yield ketone bodies in the liver • leucine (Leu) very common in proteins • lysine (Lys) Glucogenic amino acids: can be converted to glucose and glycogen • alanine (Ala) • cysteine (Cys) • glycine (Gly) • serine (Ser) • asparagine (Asn) • aspartate (Asp) • methionine (Met) • valine (Val) • arginine (Arg) • glutamine (Gln) • glutamate (Glu) • histidine (His) • proline (Pro) Mixed amino acids (both ketogenic and glucogenic): • tryptophan (Trp) • phenylalanine (Phe) • tyrosine (Tyr) • threonine (Thr) • isoleucine (Ile) Enzyme cofactors in amino acid catabolism These cofactors transfer one-carbon groups in different oxidation states: • Biotin (most oxidized: COO–) • Tetrahydrofolate (intermediate ox. state: methylene, methenyl, formyl, formimino groups, and sometimes methyl) • S-Adenosylmethionine (most reduced: methyl) Conversion of one-carbon units in tetrahydrofolate The preferred cofactor for biological methyl group transfer: adoMet This Me group is about 1,000 times more reactive than the Me group from N5-Me-tetrahydrofolate! The only other known reaction in which triphosphate is displaced from ATP occurs in the synthesis of coenzyme B12! Coenzyme B12-dependent reactions in mammals: • methionine synthase reaction • rearrangament of L-methylmalonyl-CoA to succinyl-CoA Vitamin B12 deficiency disease: metabolic folates become trapped in the N5-methyl form! Minor pathway in humans: 10-30% of Thr catabolism Oxidative cleavage pathway (the 2 C-atoms from Gly do not enter the citric acid cycle!!!) Another pathway of Gly degradation: D-amino acid oxidase: • is present at high levels in the kidney • has as primary function the detoxification of ingested D-amino acids Calcium oxalate: 75% of kidney stones! Try and Phe are precursors for biologically active molecules! The first step in Phe degradation requires the cofactor tetrahydrobiopterin: (mixed function oxidase) Mixed function oxidases: catalyze simultaneous hydroxylation of a substrate by an oxygen atom of O2 and reduction of the other oxygen atom to H2O (Urea cycle) Allosteric activator: ADP Allosteric inhibitor: GTP The primary pathway for Thr degradation in humans! • much of the catabolism of amino acids takes place in the liver • branched-chain amino acids are oxidized as fuels primarily in the muscles, adipose, kidney, and brain tissue (absent in the liver!) • branched-chain -keto acid dehydrogenase complex • pyruvate dehydrogenase complex • -ketoglutarate dehydrogenase complex similar structure, same reaction mechanism catalyze homologous reactions five cofactors: thiamine pyrophosphate FAD NAD lipoate coenzyme A inactive enzyme complex = phosphorylated form! (when the dietary intake of branched-chain amino acids is low)