* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Amino Acid Biosynthesis
Secreted frizzled-related protein 1 wikipedia , lookup
Citric acid cycle wikipedia , lookup
Paracrine signalling wikipedia , lookup
Peptide synthesis wikipedia , lookup
Signal transduction wikipedia , lookup
Proteolysis wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Point mutation wikipedia , lookup
Biochemical cascade wikipedia , lookup
Genomic imprinting wikipedia , lookup
Genetic code wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression profiling wikipedia , lookup
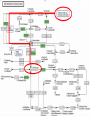
Amino Acid Biosynthesis By Laura Voss Biosynthesis vs. Metabolism • Not the same as amino acid metabolism pathways. – Synthesis of most amino acids is only one or two steps removed from another pathway or cycle. – Missing genes = metabolites not produced. Many metabolites are not essential to the cell. Biosynthesis Pathways • Valine, leucine, and isoleucine biosynthesis – Essential pathways complete • Lysine biosynthesis – Essential genes missing • Phenylalanine, tyrosine, and tryptophan biosynthesis – Essential genes missing Valine, Leucine, and Isoleucine • One missing gene: 2.3.1.182, (R)-citramalate synthase Found • (R)-citramalate synthase found by name in JGI and Manatee, not RAST – Run BLAST for amino acid sequences from JGI and Manatee on RAST • Recognizes amino acid sequences but ID’s them as isopropylmalate synthase – same COG as citramalate synthase. Lysine Lysine – missing genes • One “cluster” of genes missing • Either 1.4.1.6 or 5.1.1.7 must be present • Neither appear in any of the databases – Searching for parts of their names brings up nonspecific similar compounds, which H. utahensis might use to work around the missing genes. – Many genes don’t appear in H. marismortui either. Phenylalanine, Tyrosine, and Tryptophan Missing Genes: • 4.2.3.4 (3-dehydroquinate synthase) – Found, via name search, in all 3 databases • 5.4.99.5 (Chorismate mutase) – Found, with E.C. Number intact, in JGI and Manatee; E.C. number not in RAST. • 2.5.1.54 (3-deoxy-7-phosphoheptulonate synthase or DHAP synthase) – Not found in any database or in BLASTer search!) •EXPASY tags as DHAP synthase. •Searching for DHAP synthase in Pfam and other protein databases gives no result. •Redirects to DHAP synthetase, class I – also not found. •Also missing in H. marismortui KEGG. Conclusions and Remaining Questions: • Valine, leucine, and isoleucine complete as they appear on KEGG • Lysine does not, but alternative proteins may be present, allowing a way around missing genes. • Phenylalanine, tyrosine, and tryptophan are incomplete; no obvious substitute proteins, plus 2.5.1.54’s absense from Pfam. – Possibility: Use HMM Logo for DHAP synthetase?