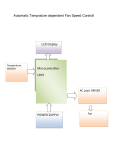
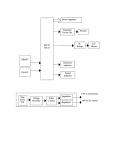
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download "Molecular Motors in Plant Cells". In: Molecular Motors
Survey
Document related concepts
Cell encapsulation wikipedia , lookup
Cell culture wikipedia , lookup
Cellular differentiation wikipedia , lookup
Extracellular matrix wikipedia , lookup
Programmed cell death wikipedia , lookup
Cell growth wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Endomembrane system wikipedia , lookup
Signal transduction wikipedia , lookup
Cytoplasmic streaming wikipedia , lookup
Transcript
18 Molecular Motors in Plant Cells A. S. N. Reddy 18.1 Introduction Molecular motors regulate diverse cellular functions including the organization and dynamics of microtubule (MT) and actin cytoskeleton, cytoplasmic streaming, cell polarity, cell growth, morphogenesis, chromosome segregation and transport of vesicles, organelles and macromolecular complexes. In eukaryotes, there are three broad families of molecular motors: the kinesins, the dyneins and the myosins. All three types of motors utilize energy from the hydrolysis of ATP to move along filamentous structures. Kinesins and dyneins move on MTs whereas the myosins translocate on actin filaments. Using biochemical, cell biological, molecular, and genetic approaches several molecular motors have been identified in plants and functions of some of these are beginning to be understood. Recent completion of genome sequence of several eukaryotes ranging from yeast, a simple unicellular eukaryote, to highly evolved multicellular organisms including humans and flowering plants has permitted comparative analysis of three families of motors in various species. Such analysis across phylogenetically divergent species has yielded some interesting insights into evolutionary and functional relationships among motor proteins from various organisms. Systematic analyses of the recently completed Arabidopsis genome sequence with the conserved motor domain of kinesins, myosins and dyneins revealed the presence of 78 molecular motors (61 kinesin-like proteins and17 myosins) in this organism. Of the two families of MT-based motors, dyneins are absent in flowering plants. Surprisingly, Arabidopsis has the largest number of kinesin-like proteins (KLPs) as compared to other multicellular organisms including humans, suggesting that the kinesin superfamily is expanded considerably in plants. Also, all plant myosins belong to two novel classes. Although the identification of molecular motors in plants is progressing rapidly due to genome and EST sequencing projects, only a few plant motors have been characterized in any detail and the functions of many these motors are not known. Nevertheless, it is becoming obvious that plants contain novel families of KLPs and myosins with novel functions and regulatory meMolecular Motors. Edited by Manfred Schliwa Copyright c 2003 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN 3-527-30594 -7 434 18.2 Microtubule-based Motors chanisms. In this article I focused primarily on plant molecular motors, their function and regulation with emphasis on the model plant, Arabidopsis. For other aspects of plant motors that are not covered here the reader is directed to other reviews (Asada and Collings, 1997, Liu and Lee, 2001, Reddy, 2001b, Yamamoto et al., 1999). 18.2 Microtubule-based Motors The motors that move on MTs belong to two different families: kinesins and KLPs that move either toward the plus-end or minus-end of the MTs, and the dyneins, which move toward the minus-end of MTs. 18.2.1 Kinesin-like Proteins Members of the kinesin superfamily, which consists of conventional kinesin and kinesin-like proteins (KLPs), are known or implicated to play important roles in many fundamental cellular and developmental processes including intracellular transport of vesicles and organelles, mitotic and meiotic spindle formation and elongation, chromosome segregation, germplasm aggregation, MT organization and dynamics and intraflagellar transport (Endow, 1999, Goldstein and Philip, 199, Hirokawa, 1998, Reddy, 2001b). A large number of KLPs have been identified in plants and animals (Berg et al., 2001, Lawrence et al., 2002, Reddy and Day, 2001a, 2001b, Siddiqui, 2002, Yamashita et al., 2000; Tab. 18.1). All members of the kinesin superfamily have a highly conserved catalytic region of about 350 amino acid residues, known as the motor domain, with ATP- and MT-binding sites. The motor domain in KLPs is located either in the N terminus, C terminus or in the middle of the protein. In addition to the motor domain, most KLPs have a stalk region that forms an alpha-helical coiled-coil region which aids in dimerization, and a highly variable tail, which is thought to interact with a specific cargo. Outside the motor domain KLPs share no sequence similarity between the members of kinesin superfamily. Although molecular motors have been implicated in a variety of cellular processes in plants including spindle function, cytokinesis, cell polarity, morphogenTable 18.1 The total number of KLPs and myosins in the completely sequenced organisms. S. cerevisiase S. pombe C. elegans D. melanogater H. sapiens A. thalaiana KLPs 6a 9a 21b 24a 45c 61a Myosins 5d 5d 17d 13d 40d 17d,e Total 11 36 37 a 14 b c 85 d 78 e Day and Reddy, 2001b; Siddiqui, 2002; Miki et al., 2001; Berg et al., 2001; Day and Reddy, 2001a 18 Molecular Motors in Plant Cells esis, organelle and vesicle transport, and cytoplasmic streaming (Asada and Collings, 1997, Lloyd, 1991, Reddy, 2001b, Williamson, 1993), until recently very little was known about their molecular identity. Using antibodies to animal kinesins, KLPs have been identified in pollen tubes of tobacco and other plants (Cai et al., 1993,; Liu et al., 1994, Tiezzi et al., 1992). Recently, Cai et al. isolated an MTbased motor (90 kD) from tobacco pollen tubes which induced MT gliding in motility assays (Cai et al., 2000). Immunolocalization studies have indicated that this motor binds to organelles associated with MTs in the cortical regions of the pollen tube, suggesting its involvement in organelle transport. Two KLPs (125 and 120 kDa) that showed plus-end motor activity in in vitro motility assays and MT-dependent ATPase and GTPase activity, were isolated from tobacco phragmoplasts (Asada and Shibaoka, 1994, Asada et al., 1991). Using various approaches cDNAs for several KLPs have been characterized from plants. Ten of these are from Arabidopsis (KatA, KatB, KatC, KatD, AtKCBP, AtPAKRP1, AtPAKRP2, AtMKRP1, AtMKRP2, AtNCAK1) (Itoh et al., 2001, Lee and Liu, 2000, Lee et al., 2001, Mitsui et al., 1996, Mitsui et al., 1994, 1993; Nishihama et al., 2002, Reddy et al., 1996b, Tamura et al., 1999), 15 from tobacco (TKRP125, KCBP, NCAK1, NCAK2, and TBK1 to TBK11) (Asada et al., 1997, Matsui et al., 2001, Nishihama et al., 2002, Wang et al., 1996), two from carrot (KRP120-1 and KRP120-2 (Barroso et al., 2000) and one from potato (KCBP). A kat gene family (katA, katB, katC, katD, and katE) encoding KLPs in Arabidopsis thaliana was characterized using primers corresponding to conserved regions of the kinesin motor domain (Mitsui et al., 1993, 1994, 1996, Tamura et al., 1999). Using a similar approach, 12 different KLPs that belong to seven subfamilies have been isolated from tobacco BY-2 cells (Matsui et al., 2001). It is interesting that in tobacco BY-2 cells alone, 15 different KLPs that belong to different subfamilies are expressed (Asada et al., 1997, Bowser and Reddy, 1997, Matsui et al., 2001, Nishihama et al., 2002). Using an indirect assay, KatA (89 kD) protein has been shown to be a minus-end-directed motor and a similar protein has been found in carrot and tobacco (Liu et al., 1996). The central region of KatD shares sequence similarity to the motor domain of kinesin and is followed by about 240 residues at the C-terminus. Phylogenetic analysis with the KatD motor domain indicates that it belongs to the C-terminal family despite the fact that it has a 240-amino acid stretch following the motor domain (Reddy and Day, 2001b, Tamura et al., 1999). The amino-terminal region of KatD showed sequence similarity to calponin homology (CH) domain. KatD is a flower-specific motor protein suggesting that it may function in transport or cytoskeleton organization in pollen. Asada et al. (1997) isolated a cDNA encoding the 125-kDa polypeptide (TKRP125, tobacco kinesin-related polypeptide of 125 kDa). It is an N-terminal KLP with strong similarity to members of the BimC (blocked in mitosis) subfamily. Two other members of BimC class KLPs (DcKRP120-1 and DcKRP120-2) were isolated from carrot suspension cells (Barroso et al., 2000). DcKRP120-1 is a homolog of TKRP125. In Arabidopsis there seems to be at least three TKRP-like proteins. Recently, two phragmoplast-associated kinesin-related proteins (AtPAKRP1 and AtPAKRP2) have been isolated from Arabidopsis and characterized (Lee and Liu, 2000, Lee et al., 2001). 435 436 18.2 Microtubule-based Motors AtPAKRP1 is an N-terminal KLP and is most closely related to XKlp2, an ungrouped KLP from Xenopus laevis. AtPAKRP2 is also an N-terminal motor with no known homologs in non-plant systems. In phylogenetic analysis, AtPAKRP2 does not group with any of the existing families of KLPs. These motors have distinct functions in cytokinesis (Lee et al., 2001). KCBP (kinesin-like calmodulin-binding protein), a novel KLP with a C-terminal motor domain was isolated from a number of plants as a calmodulin-binding protein (Abdel-Ghany et al., 2000, Abdel-Ghany and Reddy, 2000, Reddy et al., 1996a, 1996b, Wang et al., 1996). Although KCBP, like other kinesin-like proteins, contains three distinct regions (a motor domain, a coiled-coil stalk and a tail), it has two features that make it unique among members of the kinesin superfamily in eukaryotes. These include (i) a calmodulin-binding domain adjacent to the motor domain at the C-terminus, and (ii) a myosin tail homology and talin-like region in the N-terminal tail (Reddy and Reddy, 1999). KCBP binds calmodulin in a calcium-dependent manner at physiological calcium concentrations (Reddy et al., 1996b, 1999). KCBP is a minus-end directed MT motor (Song et al., 1997) and has two MT binding domains, one located at the C terminus and the second one located at the N terminus (Narasimhulu and Reddy, 1998). Unlike the MT binding domain in the C terminus, the N-terminal region of KCBP binds MTs both in the presence and absence of ATP, indicating that the MT binding domain in the N terminus is insensitive to ATP. KCBP is highly conserved in phylogenetically divergent plant species including dicots, monocots, gymnosperms and algae (S. E. Abdel-Ghany and A. S. N. Reddy, unpublished data). Recently, a calmodulin-binding C-terminal kinesin (kinesin C) was cloned from sea urchin (Rogers et al., 1999). The calmodulin-binding domain of kinesin C shared 35 % sequence identity with the calmodulin-binding domain in KCBP. The existence of calmodulin-binding KLPs in both plants and sea urchin suggests that the origin of this group of KLPs pre-dates the divergence of plants and animals from a common ancestor which is believed to have occurred about 1.5 billion years ago (Wang et al., 1999). If this was the case, there must have been insertion of some domains such as MyTH4 and talin-like regions in the tail of KCBP (or deletion of these domains in kinesin C) to acquire functional specialization of these KLPs. Alternatively, calmodulin-binding KLPs may have evolved independently in plant and animals after they diverged from a common ancestor. The amino-terminal tail and stalk regions of KCBPs from different plant systems are highly conserved and contain myosin tail homology (MyTH4) and talin-like regions that are not present in kinesin C. In phylogenetic analysis of KLPs KCBPs and kinesin C are grouped with other known C-terminal KLPs. However, Arabidopsis KCBP together with its orthologs from other plants constitute a distinct group within the C-terminal subfamily of motors. Recently, two Arabidopsis KLPs (AtMKRP1 and AtMKRP2) with mitochondrial targeting signals were found to be targeted to mitochondria, suggesting that these motors might function in this organelle (Itoh et al., 2001). AtMKRP1 and AtMKRP2 belong to ungrouped KLPs (Reddy and Day, 2001b). What functions these MKRPs perform in mitochondria remains to be seen. There are other Arabi- 18 Molecular Motors in Plant Cells dopsis KLPs with targeting signals to other organelles such as chloroplasts. It would be interesting to see if any other Arabidopsis KLPs are targeted to organelles. KLPs that are targeted to organelles have not been reported previously in the literature. Mutations in the HINKEL (HIK) gene in Arabidopsis result in defective cytokinesis. These mutants have high frequency of incomplete cell walls and multinucleate cells. The HIK gene encodes a plant-specific N-terminal KLP that plays an important role in cytokinesis (Strompen et al., 2002). In a recent report, Nishihama et al. (2002) have shown that two tobacco KLPs (NACK1 and NACK2) interact with NPK1, a mitogen-activated kinase kinase kinase, and stimulate the activity of the kinase. Orthologs of NACK1 and NACK2 have been identified in Arabidopsis and AtNACK1 was found to be identical to HIK. These KLPs are highly diverged N-terminal motors and do not group with any of the known KLP subfamilies (Reddy and Day, 2001b). The closest non-plant KLP that shares significant similarity with the motor domain of NCAKs is CENP-E. At least eight Arabidopsis KLPs are known to function in some aspect of cell division (see Section 18.4.1 on cell division). Analysis of the recently completed Arabidopsis genome sequence with the conserved motor domain of KLPs has resulted in identification of 61 KLPs in the model plant (Reddy and Day, 2001b). A corresponding cDNA or EST (Expressed Sequence Tag) was found for 37 of these KLPs (Tab. 18.2), suggesting that most KLPs in Arabidopsis are expressed and not likely to be pseudogenes. The genes encoding KLPs are distributed throughout the genome. Chromosome 3 has the highest number (18) of KLP genes. In comparison, S. cerevisiae, S. pombe, C. elegans, Drosophila, and Homo sapiens have 6, 9, 21, 24 and 45 KLPs respectively (Tab. 18.1). Surprisingly, the Arabidopsis genome contains the largest number of KLPs among all eukaryote genomes that have been sequenced. Arabidopsis has the highest percentage (0.24 %) of the total number of genes as compared to S. cerevisiae and S. pombe with 0.1 and 0.17 % respectively, C. elegans with 0.11 % and Drosophila with 0.18 %. Based on the number of KLPs in Arabidopsis, the number of cellular processes that are regulated by KLPs in plants is likely to match or exceed the number of processes controlled by animal KLPs. Only 10 of the 61 Arabidopsis KLPs have been reported in the literature (Lee and Liu, 2000, Lee et al., 2001, Itoh, 2001, Liu et al., 1996, Mitsui et al., 1993, 1994, Nishihama et al., 2002, Reddy et al., 1996b, Tamura et al., 1999) whereas the three AtKRP125 KLPs (AtKRP125a,b, and c; Tab. 18.2) show sequence similarity to a tobacco kinesin (NtKRP125) that was isolated from phragmoplasts of tobacco (Asada et al., 1997). AtKRP125b has 68 % identity with NtKRP125 over the 1000 residues they have in common. The number of known and predicted introns in KLPs range from 3 to 34 (Tab. 18.2). In the Arabidopsis genome, the number of introns ranges from 0 to 77 with an average of about five (Reddy, 2001c). More than 85 % of Arabidopsis genes have 10 or less introns while the Arabidopsis kinesin genes have an average of 16.4 with only 10 genes having 10 or less introns. The number and locations of the introns need to be verified experimentally. It is likely that the predicted sizes of 437 C-terminal C-terminal At2g36200 (AtKRP125b) At2g28620 (AtKRP125c) At3g45850 At4g05190 At4g21270 (AtKatA) At4g27180 (AtKatB) At5g54670 (AtKatC) At5g27550 At2g22610 At1g72250 At3g10310 At1g18410 At1g73860 At1g63640 At5g41310 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Internal 967 Internalc 1050 1162 * 897 1195 1068 1056 c c c c 425 Internalc Internal Internal Internal Internal c N-terminal b 754* 745* 793 777 C-terminal C-terminal 1058 1076 1056 1022 #aa N-terminal N-terminal N-terminal N-terminal At2g37420 (AtKRP125a) 1 Motor location EST/ cDNA Kinesin-like proteins in Arabidopsis. Gene code (published name) Table 18.2 19 19 17 17 16 17 18 6 15 15 8 17 21 18 22 17 # of introns a ND ND ND ND ND ND ND ND ND ND Minus ND ND ND ND ND Motility CC CC CC CC CC Other Domains ND ND ND ND ND ND ND ND ND ND CC, CH CC, CH CC CC CC, CH CC CC CC CC CC Mitotic MT arrays CC ND ND ND ND ND Cellular localization Mitsui et al, 1994. Mitsui et al, 1994. Mitsui et al., 1993; Liu et al., 1992. Asada et al., 1997. Asada et al., 1997. Asada et al., 1997; Barroso et al., 2000. Ref 438 18.2 Microtubule-based Motors N-terminal C-terminal N-terminal N-terminal At5g10470 At5g65460 At5g27950 At1g55550 At5g65930 (AtKCBP) At1g20060 At3g10180 At1g59540 At5g06670 At3g12020 At1g21730 (MKRP1) At4g39050 (MKRP2) At2g21380 21 22 23 24 25 26 27 28 29 30 31 32 33 767 At3g44730 861 1264 640 887 b b 1273 b N-terminal N-terminal N-terminal 857 1121 (1055) 909* (890) 956 997 823 459 N-terminal N-terminal 21 923* N-terminal 18 22 22 22 22 17 11 20 9 8 22 22 14 14 18 17 1259 N-terminal N-terminal N-terminal b Internal Internal c 20 At2g47500 c 19 1032* Internalc At1g09170 987 Internalc 18 At5g27000 (AtKatD) 17 ND ND ND ND ND ND ND ND Minus ND ND ND ND ND ND ND ND ND Mitochondria Mitochondria ND ND ND ND ND PPB, spindle, Spindle poles, Phragmoplast ND ND ND ND ND ND ND ND CC CC CC CC CC CC CC CC, MyTH4 Talin-like, CBD, PEST CC CC CC CC CC CC, CH CC, CH CC, CH Itoh et al., 2001. Itoh et al., 2001. Reddy et al., 1996. Tamura et al., 1999. 18 Molecular Motors in Plant Cells 439 At4g24170 At3g16630 At3g16060 At5g02370 At1g18550 At3g49650 At5g23910 At3g50240 At5g47820 41 42 43 44 45 46 47 48 49 N-terminal N-terminal N-terminal N-terminal 1032 1075 665 813 22 21 17 14 3 N-terminal 703 10 * 12 12 14 9 13 13 664 706 799 1263 968 9 11 13 11 # of introns N-terminal Internal Internal N-terminal 1087 N-terminal At5g42490 40 1037 N-terminal At5g66310 39 N-terminal At3g51150 38 581 At2g21300 37 834 932 N-terminal At4g38950 36 N-terminal 1003* (974) At3g43210 (ATNACK2) 35 N-terminal #aa N-terminal At1g18370 (HINKEL/AtNACK1) 34 Motor location EST/ cDNA (continued). Gene code (published name) Table 18.2 ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND ND Motility ND ND ND ND ND ND ND ND ND ND ND ND ND ND Interzonal MTs Phragmoplast Phragmoplast equator Cellular localization CC CC HhH1 CC CC CC CC CC CC CC CC CC CC, CC Other Domains Nishihama et al., 2002. Strompen et al., 2002; Nishihama et al., 2002. Ref 440 18.2 Microtubule-based Motors N-terminal At3g19050 At3g17360 At3g44050 At3g20150 At3g23670 At4g14150 (AtPAKRP1) At4g14330 (AtPAKRP2) 55 56 57 58 59 60 61 N-terminal 959 (869) 9 24 1292 (1294) 14 16 1103* I 20 1229 N-terminal 1268* 30 2158 N-terminal N-terminal 34 * 1070 20 16 895 2756 N-terminal N-terminal 18 * 885 11 * 24 439 1335* ND ND ND ND ND ND ND ND ND ND ND ND CC, CC CC CC, CC, CC, ARM CC, ARM CC, ARM, CC CC Punctate staining CC of Phragmoplast Spindle midzone, CC Phragmoplast ND ND ND ND ND ND ND ND ND ND Lee et al., 2001 Lee and Liu, 2000 by indirect assay; balthough the motor is N-terminal, it groups into C-terminal family; calthough the motor is internal, it groups into C-terminal family; iNCBI predicted protein has 1662 amino acids; *slight discrepancy in #aa between AtDB and MIPS predicitions; The number of amino acids in parenthesis are based on the deduced number from cDNA. EST, expressed sequence tag, CC, coiled coil; CH, calponin homology domain; MyTH4, domain present in the tail region of some myosins; Talinlike, talin-like domain found in some myosins and band 4.1 superfamily; CBD, calmodulin binding domain; PEST, motif rich in proline, glutamine, serine and threonine residues; ARM, Armadillo/beta-catenin-like repeats; HhH1, helix-hairpin-helix. (Modified from Reddy and Day, 2001a) a N-terminal At3g54870 54 N-terminal At1g12430 53 N-terminal At1g01950 52 N-terminal At3g63480 51 N-terminal At5g60930 50 18 Molecular Motors in Plant Cells 441 442 18.2 Microtubule-based Motors the proteins in Tab. 18.2 and 18.3 may change somewhat as corresponding cDNAs are characterized. In a few cases where the cDNAs were isolated recently there are some differences between the predicted and actual size of the protein due to inaccurate predictions of introns and exons (Itoh et al., 2001, Lee and Liu, 2000, Lee et al., 2001). All Arabidopsis KLPs except three have a coiled-coil region, suggesting that they may function as homo or heterodimers (Fig. 18.1 and Tab. 18.2). Interestingly, some Arabidopsis KLPs have predicted domains that are not present non-plant Table 18.3 Myosins in Arabidopsis. Name # of aa Gene code EST/ cDNA Class Domains Reference 1. At ATM 1166 AT3g19960 (ATM1)a VIII MD, CC, IQ (Knight and Kendric-Jone, 1993) 2. At ATM2 1111 1101b AT5g54280 ATM2/AtMYOS1a VIII MD, CC, IQ (Kinkema et al., 1994) 3. At VIIIA 1085 AT1g50360 VIII MD, CC, IQ 4. At VIIIB 1126 AT4g27370 VIII MD, CC, IQ 5. At MYA1 1520 1599c AT1g17580 (AtMYA1)a XI MD, CC, IQ 6. At MYA2 1505c 1515 AT5g43900 (AtMYA2)a XI MD, IQ (Kinkema and Schiefelbein, 1994) 7. At XIA 1730 AT1g04600 XI MD, CC, IQ (Kinkema et al., 1994) 8. At XIB 1519 AT1g04160 XI MD, IQ 9. At XIC 1572 AT1g08730 XI MD, CC, IQ 10. At XID 1611 AT2g33240 XI MD, CC, IQ 11. At XIE 1529 XI MD, CC, IQ XI MD, IQ XI MD, CC, IQ 12. At XIF 1556 13. At XIG 1502 AT1g54560 d AT2g31900 AT2g20290 AT4g28710 XI MD, CC, IQ 1374 AT4g33200 XI MD, CC, IQ 16. At XIJ 1242 963b 998b AT3g58160 AtMYOS3)a (AtMYA3)a XI MD, CC, IQ 17. At XIK 1544 AT5g20490 IX MD, CC, IQ 14. At XIH 1452 15. At XI-I a d b (Kinkema et al., 1994) Name as reported in the literature; Number of amino acids previously reported for partial sequence; Number of amino acids predicted by NCBI; dEdited for full-length sequence; MD, motor domain; CC, Coiled-coil region; IQ, putative calmodulin-binding motif (Modified from Reddy and Day, 2001a) c U Structural features of Arabidopsis KLPs. The deduced amino acid sequence of KLPs in the Arabidopsis database was analyzed for the presence of various domains (Reddy and Day, 2001b). The KLPs are arranged according to their grouping in the phylogenetic tree in Fig. 18.3. Characterized KLPs are indicated in Figure 18.1. MCAK/KIF2 Unc104/KIF1 Sp Chromo/KIF4 Sc BimC Ce KRP85/95 Dm KHC At MKLP1 10 8 6 4 2 0 C. Term 26 24 22 20 18 Kip3 Number of KLPs 18 Molecular Motors in Plant Cells bold. ARM, Armadillo/beta-catenin-like repeat; CBD, calmodulin-binding domain; CC, coiledcoil region; CH, Calponin homology domain; MD, motor domain; MyTH4, myosin tail homology region; Talin-like, talin-like domain; HhH1, helix-hairpin-helix domain. Bar 100 aa. Modified from Reddy and Day, 2001b. KLPs (Fig. 18.1, Tab. 18.2). Six of the Arabidopsis KLPs have a calponin homology (CH) domain, which is an actin-binding domain present in the N-termini of spectrin-like proteins. The CH domain is a protein module of approximately 110 residues found in cytoskeletal and signal transduction proteins either as two domains in tandem or as a single copy (Banuelos et al., 1998). Proteins with a tandem pair of CH domains cross-link F-actin, bundle actin or connect intermediate filaments to cytoskeleton. Proteins with a single copy are involved in signal transduction (Banuelos et al., 1998, Leinweber et al., 1999). Perhaps the KLPs containing a CH domain bind actin and are involved in signal transduction or linking of actin and MTs. Some domains that are involved in protein protein or protein DNA interactions are also present in some KLPs (Fig. 18.1 and Tab. 18.2). These include armadillo repeats, a helix-hairpin-helix DNA-binding domain. KCBP has MyTH4 and talin-like domains present in some myosins, suggesting that it has domains of both MT- and actin-based motors. Such motors may be involved in cross talk between MT and actin cytoskeleton (Reddy, 2001b). KCBP also has been shown to have a calmodulin-binding domain (Reddy et al., 1996b). Phylogenetic analysis In non-plant systems, nine subfamilies of KLPs with some ungrouped KLPs have been identified by phylogenetic analysis using the conserved motor domain (Kim and Endow, 2000). Three of the subfamilies (KHC, KRP85/95, and Unc104/KIF1) are involved in transport (Goldstein and Philip, 1999). Members 18.2.1.1 443 444 18.2 Microtubule-based Motors from the other subfamilies (C-terminal, Kip3, MKLP1, BimC, chromokinesin/ KIF4, and MCAK/KIF4) function in various processes associated with cell division (Goldstein and Philip, 1999). A phylogenetic analysis of the 61 Arabidopsis KLPs motor domain sequences with 113 other motor domain sequences from nonplant systems has revealed that seven of the nine families are represented in Arabidopsis (Figs 18.2 and 18.3). However, several Arabidopsis KLPs do not fall into any of the nine subfamilies and are likely to represent additional subfamilies that are unique to plants (Figs 18.2 and 18.3). Most of the Arabidopsis KLPs are more closely related to another Arabidopsis or another plant kinesin than to any other kinesin used in the comparison. The subfamilies that are involved in transport are under-represented in Arabidopsis (Fig. 18.2). There are no members of the KRP85/95 or Unc104/KIF1 subfamilies in Arabidopsis (Figs 18.2 and 18.3). No KHCs have been reported in plants. The phylogenetic tree indicates that there is possibly one KHC-type kinesin in Arabidopsis. At3g63480 falls into the KHC group with a closer relationship to KHCs found in fungi and, like fungal KHC, lacks a binding site for kinesin light chain proteins (Diefenbach et al., 1998; Reddy and Day, 2001b). One Arabidopsis kinesin groups with the MKLP1 subfamily (Fig. 18.3). Two internal motor Arabidopsis KLPs grouped with the MCAK/KIF2 subfamily (also called the internal family) whose members are involved in vesicle transport, chromosome movement and MT catastrophe (Goldstein and Philip, 1999). Three Arabidopsis KLPs fall into a group with Kip3 subfamily members in which ScKip3 is involved in nuclear movement (DeZwaan et al., 1997). Three Arabidopsis KLPs form a branch off of the chromokinesin/KIF4 subfamily members, some of which are involved in vesicle transport (HsKIF) and spindle organization and chromosome positioning (Xlklp1) (Goldstein and Philip, 1999). Arabidopsis has many KLPs (24 out of 61) that do not fall into any of the known groups (Figs 18.2 and 18.3). Members of BimC subfamily, which are N-terminal plus-end motors, are present in all five eukaryotic organisms that have been sequenced. The three NtKRP125-like Arabidopsis KLPs (AtKRP125a, b, and c) are grouped with the tobacco homolog in the BimC subfamily which are involved in cross-linking and anti-parallel sliding of MTs (Goldstein and Philip, 1999). Twenty-one Arabidopsis KLPs were grouped into the C-terminal subfamily. This is an unusually large number compared to the other organisms. It is also unusual in that 11 KLPs in this group have internal motors and five have N-terminal motors. The internal KLPs have a motor domain that is closer to the C-terminus than the N-terminus but each has some sequence C-terminal to the motor domain and could be called internal depending on the parameters used to define an internal motor. It will be interesting to find out the direction of movement of these KLPs whose motor domains are either N-terminal or internal but are most closely related to the C-terminal subfamily. KLPs in the C-terminal subfamily translocate toward the minus-end of MTs (Woehlke and Schliwa, 2000) and have a conserved sequence at the neck/motor core junction (Endow and Waligora, 1998). It was reported that the residues G and N residues at the neck/motor core junction are necessary for minus-end directed movement (Endow and Waligora, 1998). Analysis of the neck/motor core junction of the 21 C-terminal class Arabidopsis KLPs showed con- 18 Molecular Motors in Plant Cells Strict BimC C-Terminal MKLP1 AnBIMC ScCIN8 ScKIP1 SpoCUT7 BmKRP DmKLP61F CeF23B12.8 HsKSP XlEg5 XlEg52 NTKRP125 AtKRP125b At3g45850 AtKRP125a AtKRP125c AnKLPA SpoKLP2 SpoPKL1 ScKAR3 DmNCD CgCHO2 MmKIFC1 HsCHO2 XlCTK2 At4g05190 AtKATA AtKATB AtKATC CeKLP HsKIFC3 MsFKIF2 MmKIFC2 XlCTK1 At5g27550 At2g22610 At1g72250 At3g10310 At1g18410 At1g73860 At1g63640 At5g41310 AtKATD At1g09170 At2g47500 At3g44730 CeC41G7.2 CeMO1E11 CeWO2B12 At5g10470 At5g65460 At5g27950 At1g55550 NtKCBP StKCBP AtKCBP SpKinesinC CeMO3D4.1 CgCHO1 HsMKLP1 DmPACKLP MmKlp174 At1g20060 CrKLP1 ScKIP2 HsCENPE At3g10180 At1g59540 UmKin1 At5g06670 At3g12020 AtMKRP1 AtMKRP2 At2g21380 AtHINKEL/NACK1 Figure 18.2. The number of KLPs in each family in different organisms. C.Term, C-Terminal motor; Chromo/KIF4, chromokinesin/KIF4; U, ungrouped. At, Arabidopsis thaliana, Dm, Dro- KRP85/95 AtNACK2 At4g38950 At2g21300 At3g51150 At5g66310 At5g42490 At4g24170 CeC06G3.2 CeLC33H5 CeF20C5 HsKIF3B MmKIF3B XlKlp3 sophila melanogaster; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. Adapted from Reddy and Day, 2001b. servation of these residues in most of the C-terminal Arabidopsis KLPs (Reddy and Day, 2001b). Several Arabidopsis KLPs show some similarity to other ungrouped KLPs (Figs 18.2 and 18.3). The ungrouped centromeric proteins (HsCENPE and UmKin1) cluster with seven Arabidopsis KLPs. CENP-E binds to the kinetochore throughout mitosis and to MTs of the spindle mid-zone during late stages of mitosis (Goldstein and Philip, 1999). One Arabidopsis kinesin is paired with HsKid, 445 446 18.2 Microtubule-based Motors Unc104/KIF1 MCAK/KIF2 KIP3 Chromo/KIF4 KHC Phylogenetic tree of all Arabidopsis KLPs along with113 non-plant KLPs. The tree was generated using the motor domain of KLPs as described (Reddy and Day 2001a). Vertical dashes indicate ungrouped KLPs (light dashes, those grouped with other KLPs; bold dashes, exclusively Arabidopsis KLPs). The Arabidopsis KLPs are in bold. KLPs from the following organisms were used: An, Aspergillus nidulans; Bm, Bombyx mori; Ce, Caenorhabditis elegans; Cf, Cylindrotheca fusiformis; Cg, Cricetulus griseus; Cr, Chlamydomonas rheinhardtii; Dd, Figure 18.3. KRP85/95 HsKIF3C MmKIF3C RnKIF3C SpKIN95 CrFLA10 HsKIF3 MmKIF3A SpKIN2A DmKlp68D CeOSM3 CeF56E3 DmKlp73 CeUNC104 HsATSV MmKIF1A MmKIF1B RnKIF1B HsbKIF1C RnKIF1D CeLR144 Dm38B HsCMKRP CeK11d9.C CgMCAK RnKRP2 HsMCAK XlKCM1 HsKin2 MmKIF2 MmKIF2C XlKCM2 LmKIN At3g16630 At3g16060 CfDSK1 HsKid At5g02370 DmKlp67A At1g18550 ScKIP3 ScVII607 SpoBC2F12.13 SpoBC649.01C At3g49650 At5g23910 CeTO1G1 Ggchrkin HsKIF4 MmKIF4 Xlklp1 At3g50240 At5g47820 At5g60930 Dmklp3A CeY43F4B CeKHC DmKHC LpKHC HsKHC MmKHCS MmKHCx HsnKHC HsxKHC MmKIF5c MmKHC SpKHC NcKHC NhKin1 UmKin2 SyKin1 At3g63480 DdK7 CeLF22F4 DmNOD At1g01950 At1g12430 At3g54870 LcKIN Xlklp2 At3g19050 At3g17360 At3g44050 At3g20150 At3g23670 AtPAKRP1 AtPAKRP2 ScSMY1 Dictyostelium discoideum; Dm, Drosophila melanogaster; Gg, Gallus gallus; Hs, Homo sapiens; Lc, Leishmania chagasi; Lm, Leishmania major; Lp, Loligo pealii; Mm, Mus musculus; Ms, Morone saxatilis; Nc, Neurospora crassa; Nh, Nectria haematococca; N, Nicotiana tabacum; St, Solanum tuberosum; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Sp, Strongylocentrotus purpuratus; Spo, Schizosaccharomyces pombe; Sr, Syncephalastrum racemosum; Um, Ustilago maydis; Xl, Xenopus laevis. Modified from Reddy and Day 2001b. 18 Molecular Motors in Plant Cells an ungrouped kinesin. HsKid is a kinesin-like DNA-binding protein that is involved in spindle formation and the movements of chromosomes during mitosis and meiosis (Goldstein and Philip, 1999). Arabidopsis PAKRP1 along with five other Arabidopsis KLPs are grouped with XlKlp2. XlKlp2 is required for centrosome separation and maintenance of spindle bipolarity (Boleti et al., 1996) whereas PAKRP1 associates with the phragmoplast (Lee and Liu, 2000) and is expected to function in cytokinesis. Three Arabidopsis KLPs form a subgroup separate from any other kinesin but share a branch with CeLF22F4 and DmNOD (Fig. 18.3). Eight other Arabidopsis KLPs form a subgroup separate from KLPs of any other organism. In many cases a group of Arabidopsis KLPs forms a separate branch within the major subgroup in which they fall. The large number of KLPs in plants is expected to control many diverse cellular processes including some plant-specific functions. There are many MT-associated processes that are unique to plants (Lloyd and Hussey, 2001, Reddy, 2001b). For example, during cell division in plants several plant-specific MT arrays such as the preprophase band and the phragmoplast are formed that are important in determining the future location of the cell wall and cell wall formation, respectively (Lloyd and Hussey, 2001). These unique processes are likely to require additional plant-specific motors. In addition, centrosomes play an important role in MT organization in animals whereas plants have no well-defined centrosomes. Hence, MT organization and dynamics in plants may also require additional MT motors. The presence of unique organelles such as chloroplasts and MT-dependent processes such as cell wall synthesis may also warrant additional motors. Also, cell to cell transport of macromolecules such as RNA and viruses through plasmodesmata may require MT-based motors (Reddy, 2001b). It is very likely that plant KLPs may participate in functions other than transport, and MT dynamics and organization. Recently, it has been shown that two plant KLPs function as activators of a protein kinase (Nishihama et al., 2002). It appears that flowering plants do not have dyneins. Hence, some KLPs may be performing functions that are carried out by dyneins in other systems. 18.2.2 Dyneins Dyneins move along MTs toward the minus ends. The motor domain in dyneins (Z 1000 amino acids) is much longer than in kinesins (Z 340 amino acids) and does not share sequence similarity with the kinesin motor domain. Cytoplasmic dynein is a large multi-subunit complex consisting of two heavy chains (Z 530 kDa each), three intermediate chains (74 kDa), and four light intermediate chains (Z 55 kDa) (Hirokawa, 1998). The central and C-terminal regions of dynein heavy chain, which are predicted to form a globular structure, interact with MTs and contain motor activity whereas the N-terminal region is thought to bind cargo. The intermediate and light intermediate chains of cytoplasmic dynein associate with a protein complex called dynactin which is composed of 10 subunits (Hirokawa, 1998). Although there is some immunological and biochemical data in- 447 448 18.3 Actin-based Motors dicating the presence of dynein heavy chains in plants (Moscatelli et al., 1995), sequences that are similar to dynein and dynactin subunits have not been found in the Arabidopsis genome or plant EST databases (Lawrence et al., 2001), suggesting that they may be absent in flowering plants. The functions performed by dyneins in animals may be carried out by the expanded family members of the kinesin superfamily. Alternatively, it is possible that plant dyneins have diverged considerably, hence are not recognizable by the current search programs. 18.3 Actin-based Motors 18.3.1 Myosins Myosins, a diverse group of actin-based molecular motors, perform a broad array of cellular functions. A large number of myosins have been described in many eukaryotes (Berg et al., 2001, Reddy and Day, 2001a). Most of these are identified based on sequence similarity with the myosin motor domain. All myosin heavy chains have three common domains: a highly conserved motor domain (Z 850 amino acids) located at the amino-terminus in almost all myosins, which is followed by a neck region that binds to calmodulin or calmodulin-related proteins and a C-terminal tail domain. The calmodulin-binding domain ‘IQ motif’ in these myosins has a consensus sequence ‘IQXXXRGXXXR’ (I, isoleucine; Q, glutamine; R, arginine; G, glycine) (Wolenski, 1995). This motif is present in one to seven repeats depending on the type of myosin (Mermall et al., 1998). The only myosin that does not contain an ‘IQ motif’ is myosin XIV. The tail region of different myosins varies considerably in length and structure. The tail is characteristic to each myosin with interesting domains that are also found in non-motor proteins. The number of myosins in an organism varies among organisms. However, not all types of myosins have been identified in a single organism (Berg et al., 2001, Reddy and Day, 2001a). Although there is compelling indirect evidence for the role of actin and actinbased motors in various transport processes in plants, little is known about specific plant myosins that perform these functions. Myosins have been identified in plants both biochemically (Heslop-Harrison and Heslop-Harrison, 1989, Miller et al., 1995, Yokota et al., 1995, 1999b, Yokota and Shimmen, 1994, 1999) and at the molecular level (Kinkema and Schiefelbein, 1994, Kinkema et al., 1994, Knight and Kendrick-Jones, 1993). Immunological detection of myosins using animal myosin antibodies identified proteins of various sizes from different plants (Parke et al., 1996, Qiao et al., 1989, Tang et al., 1989). Immunofluorescence studies localized myosin to the surface of organelles, and the vegetative nuclei and generative cells in pollen grains and tubes (Heslop-Harrison and Heslop-Harrison, 1989), to the active streaming lanes and cortical surface in pollen tubes (Miller et al., 1995) and more recently to plasmodesmata in root tissues (Radford and White, 1998, Reichelt et al., 1999). Motility assays (Kohno et al., 1991) and ATPase assays 18 Molecular Motors in Plant Cells (Kohno et al., 1991, Ma and Yen, 1989; Vahey et al., 1982) using myosin-like proteins isolated from plants have also demonstrated the presence of myosins in plants. Using PCR-based approaches, a few myosins have been cloned from Arabidopsis and other plants (Kashiyama et al., 2000, Kinkema and Schiefelbein, 1994, Kinkema et al., 1994, Knight and Kendrick-Jones, 1993, Liu et al., 2001, Moepps et al., 1993). A 170-kDa myosin has been purified from lily pollen and the antibodies against this myosin stained particles of various sizes in the apical cytoplasm of lily and tobacco pollen tubes (Kohno et al., 1990, 1991, 1992, Yokota and Shimmen, 1994). These reports indicate that several myosins are likely to be present in pollen tubes. The 170- and 175-kDa myosins that translocate F-actin at a velocity of about 9 and 3 4 mm s 1, respectively, were found to be associated with calmodulin (Yokota et al., 1999a, 1999b). A myosin II-like protein was identified in Nitella by Kato et al. (Kato and Tonomura, 1977) and a myosin was purified from Chara (Yamamoto et al., 1994, 1995). In in vitro motility assays, the characean myosin translocated actin at 50 mm s 1. Rotary-shadowed images of the purified myosin showed two heads that are comparable to heads of myosin II in shape and size (Yamamoto et al., 1995). In Arabidopsis, the expression of several myosins in a given cell type implies that actin-based motors are involved in a wide range of cellular functions. By analyzing the Arabidopsis genome sequence with the conserved motor domain of myosins, a total of 17 myosins were identified in Arabidopsis (Reddy and Day, 2001b). Table 18.3 lists the myosins by name as given in the phylogenetic tree by Hodge and Cope (2000) and as assigned by Reddy and Day (2001a). In comparison, S. cerevisiae, Schizosaacharomyces pombe, C. elegans, D. melanogaster and H. sapiens have 5, 5, 17,13 and 40 myosins respectively. Five of the 17 Arabidopsis myosins have been reported in the literature (Kashiyama et al., 2000, Kinkema and Schiefelbein, 1994, Kinkema et al., 1994, Knight and Kendrick-Jones, 1993). All Arabidopsis myosins have three to six putative calmodulin-binding ‘IQ’ motifs (Fig. 18.4). The IQ domains usually follow right after the motor domain but are separated slightly from the motor domain in At XID, At XI-I, and At XIK. There are three or four IQ domains in class VIII myosins and five or six in class XI except for At XIK which has only four (Fig. 18.4). There are coiled-coil domains in all the myosins that differ in length and number. They often follow directly after the IQ domains but in some cases there is intervening sequence. Based on the presence of the coiled-coil domains, it is likely that the Arabidopsis myosins either form dimers or interact with other proteins (Cope et al., 2000). The class XI myosins are much longer than the class VIII myosins with the difference being in the length of the Cterminal region following the conserved domains found in myosins. Myosins containing IQ domains are typically calmodulin-sensitive. However, the interaction with, or regulation by calmodulin has not yet been demonstrated for Arabidopsis myosins. The information on myosin light chains is limited in plants. Recently, Yokota and his colleagues (Yokota et al., 1999a, Yokota and Shimmen, 1994, 1999) have purified two myosins from plants and demonstrated that calmodulin associates with the purified myosin and regulates its motor activity. Since the genes encoding these proteins are not cloned the structural features of these myosins are 449 450 18.3 Actin-based Motors 1. At ATM 3. At VIIIA 2. At ATM2 4. At VIIIB 13. At XIG 14. At XIH 6. At MYA2 8. At XIB 10. At XID 7. At XIA 12. At XIF 9. At XIC 11. At XIE 16. At XIJ 5. At MYA2 15. At XI-I 17. At XIK Motor Domain IQ Motif Schematic diagram showing various predicted domains in Arabidopsis myosins. The Arabidopsis genome database was searched with the conserved motor domain of a myosin to identify Arabidopsis myosins. The deduced amino acid sequence of Arabidopsis myosins Figure 18.4. Coiled-coil Domain was analyzed for the presence of various domains (Reddy and Day, 2001a). The numbers refer to the numbers in Tab. 18.1. The first four myosins are in class VIII and the following 13 are in class XI. The bar represents 100 amino acids. Adapted from Reddy and Day, 2001a. not known. Besides the motor, IQ and coiled-coil domains, several other domains have been identified in myosins from classes other than the plant classes VIII and XI. These include SH3 domains (Src homology 3 domains), ankyrin repeats, MYTH4 (domain of unknown function found in a few classes of myosins), zincbinding domain, plecstrin homology, FERM/talin (band 4.1/ezrin/radixin/moesin), GPA-rich domains and a protein kinase domain (Berg et al., 2001). Plant myosins do not have any of these other domains. 18 Molecular Motors in Plant Cells 18.3.2 Phylogenetic analysis Phylogenetic analysis using the conserved motor domain grouped all known myosins into 18 distinct classes (Burge and Rogers, 2000). Phylogenetic analysis of the Arabidopsis myosins’ motor domain with non-plant and plant myosins revealed that all the Arabidopsis myosins and other plant myosins fall into only two groups, class VIII and class XI (Fig. 18.5). Four Arabidopsis myosins are in class VIII and 13 in class XI. These groups contain exclusively plant or algal myosins with no animal or fungal myosins, suggesting that plants have a unique set of myosins (Fig. 18.5). The motor domain in all cases is in the N-terminal region (Fig. 18.4). The motor domain starts at about 50 55 residues for the class XI myosins while the class VIII myosins have a longer N-terminal region prior to the motor domain (99 159 residues). An algal (Chara corallina) myosin, Cc ccm, does group with the plant class XI myosins but is on a separate branch from any other vlass XI myosin (Fig. 18.5). A phylogenetic tree that was constructed using the full-length sequences of Arabidopsis and other plant myosins showed similar grouping of plant myosins (Reddy and Day, 2001a). Among the class XI myosins the similarity ranges from 40 85 % (full length) and 61 91 %(motor domain). The similarity between the class VIII myosins ranges from 50 83 % (full length) and 64 92 % (motor domain). When class VIII myosins are compared to class XI myosins the similarity only ranges from 22 29 % (full-length) and 35 42 % (motor domain). Myosins have 131 highly conserved residues spread throughout the motor domain that define a core consensus sequence (Cope et al., 2000). Comparison of an alignment of Arabidopsis myosin motor domains to these conserved sequences shows a great deal of conservation among them (Reddy and Day, 2001a), suggesting that they are capable of motor function. 18.4 Cellular Roles of Motors 18.4.1 Cell Division Although many of the dynamic processes of cell division such as formation of the spindle, congression of chromosomes at the equatorial region and migration of chromatids during cell division are common to both animals and plants, there are significant differences between plant and animal cell division. For example, the dividing plant cells organize their cytoskeletal elements into unique structures that are not found in non-plant systems. Just prior to prophase, plant cortical MTs rearrange to form a band of MTs called the pre-prophase band (PPB) (Goddard et al., 1994, Gunning and Wick, 1985), which accurately predicts the future location of the cell plate. Another distinctive feature of plant mitosis is the formation of a bipolar spindle in the absence of centrosomes (Franklin and Cande, 1999, Smir- 451 452 18.4 Cellular Roles of Motors 100 100 82 64 100 100 100 74 100 100 100 69 65 82 90 90 87 100 100 74 100 100 96 100 90 100 55 64 100 100 100 100 100 100 99 100 50 99 100 56 54 100 96 100 100 Phylogenetic analysis of Arabidopsis myosins. A phylogenetic tree was generated as described earlier (Reddy and Day, 2001a). The myosin groups, as defined by Hodge and Cope (2000) and Yamashita et al (2000), are identified on the left in roman numerals. The Arabidopsis myosins are in bold. Myosins from the following organisms were used: Ac, Acanthamoeba castellani; Acl, Acetabularia cliftoni; Cc, Chara corallina, Ha, Helianthus annuus; Figure 18.5. At ATM At VIIIA Ha hamy1 Zm ZMM3 At ATM2 At VIIIB Ha hamy3 At MYA1 At XIC At XIE At XIJ AT XIK Ha hamy2 Ha hamy5 Zm MYO1 At MYA2 At XIB Ha hamy4 At XIG At XIH At XIA At XID At XIF At XI-I Cc ccm Acl myo1 Acl myo2 Ac HMWMI Bt X Mm X Ce HUM-2 Mm Dilute Rn myr6 Sc Myo2 Ce HUM-3 Dm 95F Mm Waltzer Ce HUM4 En csmA Pg csm1 Ce HUM-6 Dm 35B Hs UsherIb Ce HUM-7 Hs IXa Mm IXb Rn myrV Ce IA Dm IB Hs Ib Sc MYO3 Dd IA Ce IIB Dm IIB Dd II Hs nmIIA Sc MYOI Dm PDZA Hs MysPDZ Dd MyoI Dd MyoJ Dd myoM Dm NinaC Lp III Hs XV Mm XV Pf PfM-A Tg myoA Rn XVI VIII XI XIII IV X V VI XII XVII VII IX I II XVIII III XV XIV XVI Zm, Zea mays; Bt, Bos taurus; Mm, Mus musculus; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Rn, Rattus norvegicus; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Dd, Dictyostelium discoideum; Lp, Limulus polyphemus; En, Emericella nidulans; Pg, Pyricularia grisea; Pf, Plasmodium falciparum; and Tg, Toxoplasma gondii. The numbers at the branches indicate the number of times the dichotomy was supported out of 100 bootstrap tries. 18 Molecular Motors in Plant Cells nova and Bajer, 1992). The structures involved in the organization of MTs are the centrosomes in animal cells and the spindle pole body (SPB) in fungal cells, which are lacking in plant cells (Smirnova and Bajer, 1992). Finally, cytokinesis, the process that produces two daughter cells following the completion of nuclear division, in plants is accomplished quite differently from animals. Cytokinesis in plant cells occurs via the formation of a polysaccharide cell plate, which expands from the center to the periphery (Staehelin and Hepler, 1996, Sylvester, 2000). The phragmoplast, a structure composed of two disks of parallel MTs with their plus ends toward the equatorial zone and actin filaments (Euteneur et al., 1982, Hepler and Jackson, 1968, Staehelin and Hepler, 1996, Wick, 1991), is involved in cell plate formation. Vesicles carrying the cell plate materials are transported to the equatorial zone of the phragmoplast (Samuels et al., 1995, Staehelin and Hepler, 1996, Yasuhara et al., 1993). The association of vesicles with phragmoplast MTs indicate that MT-based motors are likely to be involved in transporting the vesicles to the cell plate (Otegui and Staehelin, 2000, Otegui et al., 2001, Samuels et al., 1995). The fusion of vesicles in the cell plate is mediated by phragmoplastin, a dynamin homolog, in plants (Gu and Verma, 1996, 1997, Samuels et al., 1995). The plant spindle, which is formed in the absence of a well-defined centrosome, and several unique plant-specific MT arrays such as the pre-prophase band and the phragmoplast (Lloyd and Hussey, 2001) suggest that plants are likely to contain additional plant-specific KLPs that are not present in animal cells. In animals, six subfamilies of KLPs have been shown to be involved in some aspect of cell division (Goldstein and Philip, 1999, Reddy, 2001b). Several recent reports on KLPs expression and immuunolocalization indicate that at least seven plant KLPs (Kat A, KatB/C, TKRP125, AtKCBP, AtPAKRP1, AtPAKRP2, DcKRP120-2, HINKEL/AtNACK1) that belong to different families have mitotic function (Asada, 1996, Barroso et al., 2000, Bowser and Reddy, 1997, Lee and Liu, 2000, Lee et al., 2001, Liu et al., 1996, Nishihama et al., 2002, Smirnova et al., 1998, Strompen et al., 2002). Some KLPs are expressed in a cell cycle-dependent manner. TKRP125, a member of the BimC subfamily and NCAK from tobacco and two KLPs from Arabidopsis (KCBP, KatB/C) show a high level of expression in M-phase of the cell cycle (Asada, 1996, Bowser and Reddy, 1997, Mitsui et al., 1996, Nishihama et al., 2002). DcKRP120-1 and DcKRP120-2 are not detectable in non-dividing cells (Barroso et al., 2000). TKRP125 localizes to the cortical MTs in the S phase, along MTs of pre-prophase band (PPB) and perinuclear MTs in prophase, the equatorial plane of spindle MTs in metaphase and anaphase, and the phragmoplast MTs in telophase and cytokinesis. A carrot homolog of TKRP125 (DcKRP120-1) also localizes to the cortical MTs, the pre-prophase band, spindle and the phragmoplast (Barroso et al., 2000). Antibodies to another member of the BimC subfamily (DcKRP120-2) stained the spindle and phragmoplast, predominantly in the mid-line of the phragmoplast where plus ends of the MTs overlap (Barroso et al., 2000). AtPAKRP1, a non-member of BimC, which resembles an ungrouped Xklp2 from Xenopus, also localizes to the mid-line of the phragmoplast. KatA, a C-terminal motor, localizes near the mid-zone at metaphase and anaphase and the phragmoplast in Arabidopsis and tobacco BY-2 suspension 453 454 18.4 Cellular Roles of Motors cells (Liu et al., 1996). Anti-KCBP antibodies stain the pre-prophase band, the spindle and the phragmoplast (Bowser and Reddy, 1997). The KCBP antibody did not stain the cell plate region of the phragmoplast but MT bundles on either side of the cell plate are strongly labeled (Fig. 18.6). Localization of KCBP in Haemanthus also showed localization to mitotic MTs arrays. In Haemanthus anti-KCBP staining is seen almost exclusively at the spindle poles in late anaphase (Smirnova et al., 1998). The assembly of the acentriolar spindle in plants may involve convergence of MT minus-ends leading to the formation of spindle poles (Franklin and Cande, 1999, Smirnova and Bajer, 1992). The fact that KCBP is a minus-end-directed motor (Song et al., 1997) and localizes to the spindle poles suggests that it may be involved in acentriolar spindle formation in plants. Between early and late anaphase, the localization of KCBP shifts toward the spindle pole, supporting a role for KCBP in the formation of a converging bipolar spindle (Smirnova et al., 1998, Smirnova and Bajer, 1992). Localization of minus-end (KatA and KCBP) and plus-end (TKRP125) motors in the spindle suggests their involvement in counterbalancing forces generated by plus- and minus-end motors to stabilize the spindle. Constitutive activation of KCBP during late prophase caused premature breakdown of the nuclear envelope and arrest of cells at pro-metaphase whereas activation of KCBP at late metaphase or anaphase did not effect the progression of anaphase but caused aberrant phragmoplasts formation and delayed cytokinesis (Vos et al., 2000). This study suggests that KCBP is differentially active during the various phases of cell division. Its activity is downregulated in metaphase and telophase and upregulated in anaphase most likely by changes in cytosolic calcium level (Vos et al., 2000). Co-localization of some of the KLPs with mitotic MT arrays suggests that they may have a role in forming these arrays by bundling MTs. The motor domain of KCBP has been shown to bundle MTs (Kao et al., 2000). Numerous forces are at play in the development, maintenance, and function of the phragmoplast. Golgi-derived vesicles are transported to the forming cell plate where the plus-ends of phragmoplast MTs interdigitate. It is likely that MT motors are involved in forming the phragmoplast and powering the movement of the vesicles along the MTs. Seven plant KLPs (Kat A, TKRP125, AtKCBP, AtPAKRP1, AtPAKRP2, DcKRP120-2, HINKEL/AtNACK1) including plus- and minus-end motors and some plant-specific KLPs are associated with the phragmoplast and so are expected to function in cytokinesis (Asada, 1996, Barroso et al., 2000, Bowser and Reddy, 1997, Lee and Liu, 2000, Lee et al., 2001, Liu et al., 1996, Nishihama et al., 2002, Smirnova et al., 1998, Strompen et al., 2002). The localization patterns with various KLPs differed considerably. Immunolocalization studies as well as phragmoplast MT gliding assay in the presence of TKRP antibodies indicates its involvement in the organization of MT arrays especially in phragmoplast function (Asada et al., 1997). The polarity of MTs and microfilaments in the phragmoplast (Euteneur et al., 1982, Staehelin and Hepler, 1996) indicates that plus-end motors are likely to be involved in the transport of vesicles to the cell plate. Of the seven KLPs that localize to the phragmoplast, five are N-terminal motors (TKRP125, PAKRP1, DcKRP120-2, PAKRP2, HIK/NCAK) and two are C-terminal (KatA and KCBB). TKRP125 is a plus-end motor, and the other four N-terminal 18 Molecular Motors in Plant Cells DAPI MT Triple localization of nucleus, MTs and KCBP in a cytokinetic cell of tobacco. Nuclei stained with DAPI (DAPI), MTs (MT) are localized with a monoclonal antibody to b-tubulin and KCBP (KCBP) was localized with affinitypurified anti-KCBP antibody. Secondary antibodies conjugated to FITC and Cy3 were used to detect MTs and KCBP, respectively. From Bowser and Reddy, 1997. Figure 18.6. KCBP KLPs are likely plus-end motors. One or more of these are likely to be involved in vesicle transport to the cell plate. The punctate staining pattern of PAKRP2 and its association with vesicles, suggest that it is involved in transporting vesicles to the cell plate (Lee et al., 2001). It is unlikely that KCBP and KatA play a role in vesicle transport to the cell plate as these are known to be minus-end motors and the MTs are oriented with their plus ends facing the cell plate (Euteneur et al., 1982, Liu et al., 1996, Song et al., 1997). The possible functions of minus-end motors are stabilization of the phragmoplast and/or recycling of Golgi vesicle membranes from the expanding cell plate. Functional analysis of TKRP125 indicates its role in crosslinking and sliding of anti-parallel MTs in the phragmoplast (Asada et al., 1997, Asada and Shibaoka, 1994). Immunofluorescence with AtPAKRP1 antibodies showed staining of interzonal MTs at late anaphase and later on its localization is restricted to the plus ends of interdigitating MTs in the phragmoplast (Lee and Liu, 2000). Functional studies with this KLP indicates its involvement in maintaining the integrity of phragmoplast MTs (Lee and Liu, 2000). In order for the growth of the phragmoplast to be achieved, a rapid turnover of phragmoplast MTs must occur (Hepler and Hush, 1996, Hush et al., 1994). The expansion of 455 456 18.4 Cellular Roles of Motors the phragmoplast from the center to the periphery involves disassembly of the MTs inside and assembly of MTs at the outside. Blocking of disassembly in the center by MT stabilizing drugs causes cessation of phragmoplast expansion and formation of incomplete cell walls (Nishihama and Machida, 2001, Yasuhara et al., 1993). Phragmoplast-associated KLPs may play a role in the MT assembly and disassembly associated with its growth. The loss of MTs from the center of the phragmoplast as it grows does not occur in hik mutants, suggesting that it controls directly or indirectly the dynamics of MTs in the phragmoplast (Strompen et al., 2002). HIK does not appear to be involved in either vesicle transport or organization of MTs in the phragmoplast. Studies with tobacco NCAK1, a homolog of HINKEL, showed its localization to the equatorial zone of the phragmoplast but not in the inner regions of the phragmoplast where the cell plate matures (Nishihama et al., 2002). It appears that NCAK through a protein kinase, NPK1, regulates depolymerization of MTs in the center of the phragmoplast as it expands (Nishihama et al., 2001, 2002). There is some evidence for the presence of a myosin in the phragmoplast (Parke et al., 1996), suggesting that actin-based motors may also be involved in the transport of vesicles on the phragmoplast. During cell division the anti-class VIII myosin staining remains confined to the transverse cell walls and is strongest in the newly formed cell wall (Reichelt et al., 1999). Immunogold electron microscopy showed labeling of class VIII myosin associated with the plasma membrane and plasmodesmata. These studies suggest that class VIII myosins may be involved in new cell wall formation and transport in the plasmodesmata. 18.4.2 Cell Polarity and Morphogenesis Both actin and MT cytoskeleton have been implicated in cell polarity and morphogenesis (Mathur and Chua, 2000, Mathur et al., 1999, Reddy and Day, 2000, Szymanski et al., 1999, Vidali and Hepler, 2001). The first indication that a KLP might play a role in morphogenesis came from studies with the zwichel mutant. In wild-type Arabidopsis, trichomes are unicellular with a stalk and three branches (Oppenheimer, 1998). In zwichel (zwi) mutants trichomes are abnormal with a short stalk and one or two branches depending on the severity of the allele (Fig. 18.7). Cloning of ZWICHEL has indicated that it is identical to KCBP, a calmodulin-binding KLP (Oppenheimer et al., 1997), suggesting the requirement of this motor for expansion of the stalk and branching. Using paclitaxel, a known MT stabilizer, zwichel mutants were induced to form similar growth points indicative of branch formation in normal trichomes (Mathur and Chua, 2000). Transient stabilization of MTs could compensate for KCBP/ZWI activity suggesting that it may be involved in stabilization of MTs. The two MT-binding sites of KCBP (Narasimhulu and Reddy, 1998) could be involved in stabilization or possibly a KCBP/ ZWI-interacting protein(s) could be responsible for the stabilization. Several alleles of zwi have been characterized. All zwi mutants grew normally with no apparent defects in cell division except that they contain abnormal trichomes (Krishnakumar and Oppenheimer, 1999, Oppenheimer et al., 1997). The lack of a phenotype in 18 Molecular Motors in Plant Cells Scanning electron micrographs of Arabidopsis trichomes of wild-type and KLP (zwichel) mutants. (a) Wild-type; (b) zwi; (c) zwi w2; (d) zwi9311-11. In zwi w2 most trichomes have two branches but the length of the second branch is varied. In the zwi9311-11 mutant Figure 18.7. about 40 % of trichomes are unbranched and the rest have two branches. Seeds of zwichel mutants were kindly provided by Dr David Oppenheimer and Dr Martin Hülskamp. Scale bar 100 mm. From Reddy and Day, 2000. other tissues suggests that either KCBP is non-essential or another motor with overlapping functions may substitute for ZWI function in other tissues. In Arabidopsis there are 61 KLPs including several C-terminal motors. Three extragenic suppressors (suppressor of zwichel-3; suz1, suz2 and suz3) that rescued the trichome branch number defect in a zwichel mutant have been isolated (Krishnakumar and Oppenheimer, 1999). All three suppressors were found to be allele specific, indicating direct interactions between these proteins. The suz1 zwi-3 double mutants are male sterile due to a defect in pollen germination and pollen tube growth, suggesting a role for these genes in pollen germination and tube growth (Krishnakumar and Oppenheimer, 1999). At least two proteins, a plant-specific protein kinase and a protein required for trichome branching (ANGUISTIFOLIA), have been shown to interact with KCBP/ZWI (Day et al., 2000, Folkers et al., 2002). 18.4.3 Cytoplasmic Streaming Cytoplasmic streaming in plant cells is important in intracellular transport. The force that drives the cytoplasmic streaming appears to be dependent on actin and actin-based motors (Shimmen and Yokota, 1994, Staiger and Schilwa, 1987, Williamson, 1976). Insight into the function of plant myosins in cytoplasmic streaming has been gained primarily by studies carried out with algae and pollen tubes. Characean cells exhibit a very rapid cytoplasmic streaming (Z 70 mm s 1). Also in pollen tubes, vesicles flow rapidly from the shank to the tip of the pollen tube where they fuse to support extensive tip growth of these cells (Emons et al., 1991, Heslop-Harrison and Heslop-Harrison, 1990). In addition to the bi-directional movement of vesicles, the generative cell and the nucleus move unidirectionally toward the tip (Mascarenhas, 1990, 1993). In Chara, an increase in Ca2 concentration causes cytoplasmic streaming to stop (Hayama et al., 1979). A myosin isolated from the alga Chara corallina was shown to be responsible for cytoplasmic 457 458 18.4 Cellular Roles of Motors streaming (Yamamoto et al., 1994, 1995, 1999). The myosin was cloned and found to be a class XI myosin related to the Arabidopsis MYA myosins (Kashiyama et al., 2000). Using immunofluorescence, myosin was localized to vesicles, organelles and generative cells and vegetative nuclei in grass pollen tubes (Heslop-Harrison and Heslop-Harrison, 1989). A myosin isolated from lily pollen has been shown to be responsible for cytoplasmic streaming in pollen tubes and two myosins identified in tobacco cell cultures are also thought to participate in cytoplasmic streaming (Yokota and Shimmen, 1994, Yokota et al., 1999b). Myosin has been localized to vesicles, chloroplasts and other organelles, indicating their involvement in transport of these organelles (Heslop-Harrison and Heslop-Harrison, 1989, Miller et al., 1995). The stop-and-go movements of Golgi stacks in plants also appear to be dependent on myosin (Nebenfuhr et al., 1999). However, specific myosin(s) responsible for the movement of these organelles have not been identified. Biochemical properties and the localization data of two lily pollen myosins (175 and 170 kDa) suggest that these myosins are at least partially responsible for cytoplasmic streaming, organelle transport and calcium sensitivity of cytoplasmic streaming (Yokota et al., 1999a,1999b, Yokota and Shimmen, 1999). It has been shown that the directional movement of mitochondria is dependent on the F-actin and myosin system (Van Gestel et al., 2002). Unlike in animals, the movement of peroxisomes in plants is actin based and actin polymerization appears to drive the movement of these organelles (Mathur et al., 2002). 18.4.4 Microtubule Dynamics and Organization Microtubules are highly dynamic structures with a half-life of an individual MT of about 10 min. Drugs that disrupt the dynamics of MTs have been shown to severely hamper the MT-dependent process (Reddy, 2001b). Treatment of dividing cells with colchicine leads to disappearance of the mitotic spindle, suggesting that MTs in the spindle undergo continuous polymerization and deploymerization. Taxol, in contrast to colchicine, binds to MTs and stabilizes them. Addition of taxol to dividing cells leads to arrest of dividing cells in mitosis. The dynamics of the MT cytoskeleton during cell division in plant cells have been studied extensively (Baskin and Cande, 1990, Hepler and Hush, 1996). The rapid turnover of plant MTs suggests that the transition between MT arrays and change in orientations may involve depolymerization/re-polymerization of MTs and/or movement of MTs. Dynamic instability of MTs is important for their normal function. For example, stabilization of MTs in elongating root hairs causes loss of growth directionality and promotes branching (Bibikova et al., 1999). The MTs of the phragmoplast have a t of 60 s (Hush et al., 1994). However, the mechanisms that regulate the formation and dynamic instability of MT arrays in plants are not understood. Several aspects of MT dynamics such as cross-linking, zippering, bundling, sliding, and stability of MTs is regulated by motor proteins (Goldstein and Philip, 1999). KCBP like Ncd in Drosophila can cross-link and zipper MTs to focus MTs to form spindle poles (Chandra et al., 1993, Kao et al., 2000, Matthies et al., 1996, Smirnova et al., 1998). Two dis- 18 Molecular Motors in Plant Cells tinct MT binding sites and minus-motor activity enable these proteins to perform such a function. In addition C-terminal motors have been shown to bundle MTs (Chandra et al., 1993, Kao et al., 2000). Some motors are implicated in polymerization and depolymerization of cytoskeletal elements. There is at least one example where a KLP has no motor activity but controls MT dynamics (Desai et al., 1999). Plant KLPs such as NCAK1 appear to be involved in MT dynamics (Nishihama et al., 2002, Strompen et al., 2002). Hence, the paradigm that motors bind cargo and move along cytoskeletal tracks may not explain the functions of all the motors. 18.4.5 Intercellular Transport Plant cells are connected through plasmodesmata to form a continuous symplastic network. Recent studies show that macromolecules such as RNA and protein move from cell to cell through plasmodesmata (Lazarowitz and Beachy, 1999, Lucas et al., 1995, McLean et al., 1995, 1997). The cytoskeleton and molecular motors have been implicated in this regulated transport of macromolecules through plasmodesmata (Blackman and Overall, 1998, McLean et al., 1995, White et al., 1994). The movement protein of viruses which enables the movement of viruses from cell to cell colocalizes to MTs, suggesting a role for cytoskeleton and possibly molecular motors in the transport of protein from cell to cell through plasmodesmata (Heinlein et al., 1995, Lazarowitz and Beachy, 1999, Mas and Beachy, 1999, 2000, McLean et al., 1995). These studies hint at an interesting possibility that motors may play a role in transporting macromolecules from cell to cell and influence cell-to-cell communication. Immunolocalization studies have also detected myosin associated with plasmodesmata. A recent study using an antibody to a cloned class VIII Arabidopsis myosin ATM1 (At ATM) localized this myosin to the plasmodesmata and the plasma membrane regions involved in the assembly of new cell walls (Reichelt et al., 1999). Earlier work suggested that actin was involved in regulation of plasmodesmal transport (Ding et al., 1996). Other studies using antibodies to animal myosins in root tissues of Allium cepa, Zea mays and Hordeum vulagare have also indicated the presence of myosin in the plasmodesmata (Radford and White, 1998). 18.4.6 Other functions It appears that certain plant motors may perform unexpected novel functions. For example, two plant KLPs (NCAK1 and NCAK2) have been shown to interact with NPK1, a MAP kinase kinase kinase, and stimulate its activity (Nishihama et al., 2002). Motors in plants may also be involved in regulating cell polarity, development and differentiation by transporting the proteins or mRNA to localized regions within the cell (Arn and MacDonald, 1998). There is substantial indirect evidence indicating that asymmetric localization of mRNA (e. g. ASH1 mRNA in yeast), protein (e. g. calmodulin in Drosophila photoreceptors, a MAPKKK in 459 460 18.5 Regulation of Motors tobacco BY2 cells) and RNA protein complexes in some eukaryotic cells is dependent on cytoskeletal motors (Bertrand et al., 1998, Carson et al., 1997, 1998, Long et al., 1997, Munchow et al., 1999, Nishihama et al., 2002, Oleynikov and Singer, 1998, Perhonen et al., 1998, Porter et al., 1993, 1995; Takizawa et al., 1997). 18.5 Regulation of Motors Although a large number of KLPs have been characterized from diverse organisms (www.blocks.fhcrc.org/kinesin/), the mechanisms that regulate the activity/function of MT-dependent motors is limited both in plants and animals (Reddy, 2001b, Reilein et al., 2001). 18.5.1 Calcium/Calmodulin Calcium is a key messenger in transducing many hormonal and environmental signals in plants (Reddy, 2001a). Calmodulin, a ubiquitous calcium-binding protein in eukaryotes, is highly conserved in eukaryotes and regulates many diverse cellular functions by modulating the activity of the proteins that interact with it. Upon binding calcium, calmodulin becomes activated via a conformational change and is able to regulate the activity of its target proteins (Reddy, 2001a). Cytoplasmic streaming of characean cells is regulated by the cytosolic calcium concentration (Nagai, 1993, Shimmen and Yokota, 1994, Williamson and Ashley, 1982). Elevated levels of calcium inhibit cytoplasmic streaming and the movement of organelles (Hayama et al., 1979, Kikuyama and Tazawa, 1982, Kohno and Shimmen, 1987, 1988a, 1988b, Takagi and Nagai, 1986, Tominaga et al., 1983, Williamson and Ashley, 1982). Movement of pollen tube organelles along actin cables is inhibited by calcium, suggesting that the activity of actin-based motors is sensitive to calcium levels. The mechanisms by which calcium regulates the activity of these motors is just beginning to emerge. All myosins in Arabidopsis possess two or more putative calmodulin-binding motifs, suggesting that myosins in plants are likely to be regulated by calmodulin in response to changes in intracellular calcium. Two myosin heavy chains (170 and 175 kDa) in plants associate with calmodulin, indicating that calmodulin serves as a light chain for these myosins. Yokota et al. (1999a,b) using highly purified plant myosins have provided the first evidence that calcium inhibits the myosin motor activity as well as myosin-activated ATPase activity in plants. The inhibition of myosin activity in the presence of calcium appears to be due to dissociation of calmodulin from the heavy chain since calcium inhibition of motor activity can be restored by exogenous addition of calmodulin. KCBP and recently discovered kinesin C are the only KLPs among the kinesin superfamily that bind calmodulin (Reddy et al., 1996b, Rogers et al., 1999). The binding of activated calmodulin to KCBP inhibits its interaction with MTs via the calmodulin-binding domain resulting in the inhibition of MT-dependent 18 Molecular Motors in Plant Cells ATPase activity and motility (Deavours et al., 1998, Narasimhulu et al., 1997, Narasimhulu and Reddy, 1998, Song et al., 1997). These studies strongly suggest that the binding of calmodulin to KCBP affects MT binding regions resulting in inhibition of KCBP binding to MTs and dissociation of the KCBP/MT complex. Structural studies with KCBP in free and calmodulin-bound forms are needed to verify these speculations. The calmodulin effect on KCBP suggests that activated calmodulin can act as a molecular switch to downregulate the activity of KCBP. Based on in vitro studies with KCBP it is reasonable to speculate that spatial and temporal changes in free cytosolic calcium levels in response to signals are likely to regulate KCBP activity in the cell. 18.5.2 Protein Phosphorylation The members of the BimC family have a conserved ‘BimC Box’ in which a threonine residue is phosphorylated by cdc2-kinase during cell division. Targeting of the KLP to the spindle is regulated by phosphorylation (Blangy et al., 1995, Sawin and Mitchison, 1995). TKRP125, all three members of Arabidopsis TKRP-like proteins (AtKRPa, b and c) and DcKRP120-1 and DcKRP120-2 that belong to BimC family have the conserved BimC box, suggesting that phosphorylation of these KLPs may be involved in targeting them to the right cellular location. Recently it was found that the tail region of KCBP also interacts with a protein kinase (KIPK, KCBP interacting protein kinase) in the yeast two-hybrid system (Day et al., 2000). The association of KIPK with KCBP suggests regulation of KCBP or KCBP-associated proteins by phosphorylation and/or KCBP is involved in targeting KIPK to its proper cellular location. The tobacco NPK1, a mitogen-activated protein kinase kinase kinase, NPK1, interacts with NACKs. This interaction results in activation and autophosphorylation of NPK1 and phosphorylation of NACK1 (Nishihama et al., 2002). Although the significance of NACK1 phosphorylation is not known, NPK1 phosphorylation targets it to the equatorial zone of the phragmoplasts (Nishihama et al., 2002). 18.6 Concluding Remarks The complete sequencing of several phylogenetically diverse model organisms ranging from a unicellular eukaryote to highly complex multicellular animals and a plant has allowed comparative analysis of molecular motors in eukaryotes. Plants contain a large number of motors. About 0.4 % of the predicted genes in Arabidopsis encode molecular motors. The comparative analysis provides a framework for future functional studies with plant KLPs. The number of myosins in Arabidopsis is comparable to other multicellular organisms of similar genome size. However, plant myosins are unlike myosins from any other organism except algae. Nonplant myosins contain a variety of other known domains that are lacking in plants. 461 462 References Furthermore, Arabidopsis has a surprisingly large number of KLPs among the completed eukaryotic genomes. The role of molecular motors in many plant cellular processes is beginning to be unraveled. Although the functions of most of the plant motors remain to be determined, phylogenetic analysis and known domains in these proteins are providing clues to their function which can be tested empirically. Many Arabidopsis KLPs do not fall into any known subfamilies of KLPs and several Arabidopsis KLPs are not present in other organisms and are likely to represent new plant-specific subfamilies. Analysis of expression of each motor and cellular localization, using reporter fusions coupled with studies using the loss-of-function and gain-of-function mutants, should help to elucidate the functions of these motors. Identification of the proteins that are associated with motors is also important to our understanding of their roles in plants. Knockout mutants for almost all Arabidopsis motors are already available from SIGnAL (http://signal.salk.edu/) or TMRI (http://www.tmri.org). Because of the large number of motors and likely functional redundancy or overlap with other motors it will be necessary to create not only single mutants but also double or triple mutants. Analysis of the function and regulation of plant motors will be an exciting area of research in plant cell biology and is bound to produce many surprises. Acknowledgments I thank members of my laboratory and all my collaborators for their contributions. I wish to thank Irene Day for preparing the figures and critically reading the manuscript. Research on motors in my laboratory is supported by grants from the National Science Foundation (MCB-9630782 and MCB-0079938). References Abdel-Ghany, S. E., P. Kugrens, and A. S. N. Reddy. 2000. CpKLP1: A calmodulin-binding kinesin-like protein from Cyanophora paradoxa (glaucophyta). J. Phycol. 36: 686 692. Abdel-Ghany, S. E. and A. S. N. Reddy. 2000. A novel calcium/calmodulin-regulated kinesinlike protein is highly conserved between monocots and dicots. DNA Cell Biol. 19: 567 578. Arn, E. A. and P. M. MacDonald. 1998. Motors driving mRNA localization: new insights from in vivo imaging. Cell. 95: 150 154. Asada, T. 1996. A kinesin-related motor associated with plant specific microtubule systems. Plant Cell Physiol. 37 (Suppl.): S08. Asada, T. and D. Collings. 1997. Molecular motors in higher plants. Trends Plant Sci. 2: 29 37. Asada, T., R. Kuriyama, and H. Shibaoka. 1997. TKRP125, a kinesin-related protein involved in the centrosome-independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J. Cell Sci. 110: 179 189. Asada, T., and H. Shibaoka. 1994. Isolation of polypeptides with microtubule-translocating activity from phragmoplasts of tobacco BY-2 cells. J. Cell Sci. 107: 2249 2257. Asada, T., S. Sonobe, and H. Shibaoka. 1991. Microtubule translocation in the cytokinetic apparatus of cultured tobacco cells. Nature 350: 238 241. Banuelos, S., M. Saraste, and K. D. Carugo. 1998. Structural comparisons of calponin homology domains: implications for actin binding. Structure 6: 1419–31. 18 Molecular Motors in Plant Cells Barroso, C., J. Chan, V. Allan, J. Doonan, P. Hussey, and C. Lloyd. 2000. Two kinesin-related proteins associated with the cold-stable cytoskeleton of carrot cells: characterization of a novel kinesin, DcKRP120-2. Plant J. 24: 859 868. Baskin, T. I. and W. Z. Cande. 1990. The structure and function of the mitotic spindle in flowering plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 277 315. Berg, J. S., B. C. Powell, and R. E. Cheney. 2001. A millennial myosin census. Mol. Biol. Cell. 12: 780 794. Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2: 437 445. Bibikova, T. N., E. B. Blancaflor, and S. Gilroy. 1999. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17: 657 665. Blackman, L. M. and R. L. Overall. 1998. Immunolocalization of the cytoskeleton to plasmodesmata of Chara corallina. Plant J. 14: 733 741. Blangy, A., H. A. Lane, P. d’Hérin, M. Harper, M. Kress, and E. A. Nigg. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159 1169. Boleti, H., E. Karsenti, and I. Vernos. 1996. Xklp2, a novel xenopus centrosomal kinesinlike protein required for centrosome separation during mitosis. Cell 84: 49 59. Bowser, J. and A. S. Reddy. 1997. Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J. 12: 1429 1437. Burge, H .A. and C. A. Rogers. 2000. Outdoor allergens. Environ Health Perspect. 108 (Suppl. 4): 653 659. Cai, G., A. Bartalesi, C. Del Casino, A. Moscatelli, A. Tiezzi, and M. Cresti. 1993. The kinesin-immunoreactive homologue from Nicotiana tabacum pollen tubes: Biochemical properties and subcellular localization. Planta 191: 496 506. Cai, G., S. Romagnoli, A. Moscatelli, E. Ovidi, G. Gambellini, A. Tiezzi, and M. Cresti. 2000. Identification and characterization of a novel microtubule-based motor associated with membranous organelles in tobacco pollen tubes. Plant Cell 12: 1719 1736. Carson, J. H., S. Kwon, and E. Barbarese. 1998. RNA trafficking in myelinating cells. Curr. Opin. Neurobiol. 8: 607 612. Carson, J. H., K. Worboys, K. Ainger, and E. Barbarese. 1997. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytoskeleton 38: 318 328. Chandra, R., E. D. Salmon, H. P. Erickson, A. Lockhart, and S. A. Endow. 1993. Structural and functional domains of the Drosophila ncd microtubule motor protein. J. Biol. Chem. 268: 9005 9013. Cope, M. J., J. Whisstock, and I. Rayment. 2000. Conservation within the myosin motor domain: implications for structure and function. Structure 4: 969 986. Day, I. S., C. Miller, M. Golovkin, and A. S. N. Reddy. 2000. Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J. Biol. Chem. 275: 13737 13745. Deavours, B. E., A. S. N. Reddy, and R. A. Walker. 1998. Ca2/calmodulin regulation of the Arabidopsis kinesin-like calmodulinbinding protein. Cell Motil. Cytoskeleton 40: 408 416. Desai, A., S. Verma, T. J. Mitchison, and C. E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell 96: 69 78. DeZwaan, T. M., E. Ellingson, D. Pellman, and D. M. Roof. 1997. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138: 1023 1040. Diefenbach, R. J., J. P. Mackay, P. J. Armati, and A. L. Cunningham. 1998. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 37: 16663–16670. Ding, B., M. O. Kwon, and L. Warnberg. 1996. Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 10: 157 164. Emons, A. M. C., E. S. Pierson, and J. Derksen. 1991. Cytoskeleton and intracellular movements in plant cells. In: Biotechnology: Current Progress. Edited by P. N. Cherimisimoff and L. Ferrante. Basel: Technomic, pp. 311 335. Endow, S. A. 1999. Microtubule motors in spindle and chromosome motility. Eur. J. Biochem. 262: 12 18. 463 464 References Endow, S. A. and K. W. Waligora. 1998. Determinants of kinesin motor polarity. Science 281: 1200 1202. Euteneur, U., W. T. Jackson, and J. R. McIntosh. 1982. Polarity of spindle microtubules in Haemanthus endosperm. J. Cell Biol. 94: 644 653. Folkers, U., V. Kirik, U. Schobinger, S. Falk, S. Krishnakumar, M. A. Pollock, D. G. Oppenheimer, I. Day, A. S. N. Reddy, G. Jurgens, and M. Hulskamp. 2002. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 21: 1280 1288. Franklin, A. E. and W. Z. Cande. 1999. Nuclear organization and chromosome segregation. Plant Cell. 11: 523 534. Goddard, R. H., S. M. Wick, C. D. Silflow, and D. P. Snustad. 1994. Microtubule components of the plant cell cytoskeleton. Plant Physiol. 104: 1 6. Goldstein, L. S. B. and A. V. Philip. 1999. The road less traveled: Emerging principles of kinesin motor utilization. Annu. Rev. Cell Dev. Biol. 15: 141 183. Gu, X. and D. P. Verma. 1996. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15: 695 704. Gu, X. and D. P. Verma. 1997. Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell. 9: 157 169. Gunning, B. E., and S. M. Wick. 1985. Preprophase bands, phragmoplasts, and spatial control of cytokinesis. J. Cell. Sci. Suppl. 2: 157 179. Hayama, T., T. Shimmen, and M. Tazawa. 1979. Participation of Ca2 in cessation of cytoplasmic streaming induced by membrane excitation in Characeae internodal cells. Protoplasma 99: 305 321. Heinlein, M., B. L. Epel, H. S. Pagfett, and R. N. Beachy. 1995. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science. 270: 1983 1985. Hepler, P. K. and J. M. Hush. 1996. Behavior of microtubules in living plant cells. Plant Physiol. 112: 455 461. Hepler, P. K. and W. K. Jackson. 1968. Microtubules and early stages of cell-plate formation in the endosperm of Haemanthus katherinae Baker. J. Cell Biol. 38: 437 446. Heslop-Harrison, J., and Y. Heslop-Harrison. 1989. Myosin associated with the surface of organelles, vegetative nuclei and generative cells in angiosperm pollen grains and tubes. J. Cell Sci. 94: 319 325. Heslop-Harrison, J., and Y. Heslop-Harrison. 1990. Dynamic aspects of apical zonation in the angiosperm pollen tube. Sex. Plant Reprod. 3: 187 194. Hirokawa, N. 1998. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519 526. Hodge, T., and M. J. Cope. 2000. A myosin family tree. J. Cell Sci. 113: 3353 3354. Hush, J. M., P. Wadsworth, D. A. Callaham, and P. K. Hepler. 1994. Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107: 775 784. Itoh, R., M. Fujiwara, and S. Yoshida. 2001. Kinesin-related proteins with a mitochondrial targeting signal. Plant Physiol. 127: 724 726. Kao, Y.-L., B. E. Deavours, K. K. Phelps, R. Walker, and A. S. N. Reddy. 2000. Bundling of microtubules by motor and tail domains of a kinesin-like calmodulin-binding protein from Arabidopsis: Regulation by Ca2/calmodulin. Biochem Biophys Res Commun. 267: 201 207. Kashiyama, T., N. Kimura, T. Mimura, and K. Yamamoto. 2000. Cloning and characterization of a myosin from characean alga, the fastest motor protein in the world. J Biochem (Tokyo). 127: 1065 1070. Kato, T. and Y. Tonomura. 1977. Identification of myosin in Nitella flexilis. J. Biochem. 82: 777 782. Kikuyama, M., and M. Tazawa. 1982. Ca2 ion reversibly inhibits the cytoplasmic streaming of Nitella. Protoplasma 113: 241 243. Kim, A. J. and S. A. Endow. 2000. A kinesin family tree. J. Cell Sci. 113: 3681 3682. Kinkema, M. and J. Schiefelbein. 1994. A myosin from a higher plant has structural similarities to class V myosins. J. Mol. Biol. 239: 591 597. Kinkema, M., H. Wang, and J. Schiefelbein. 1994. Molecular analysis of the myosin gene family in Arabidopsis thaliana. Plant. Mol. Biol. 26: 1139 1153. Knight, A. E. and J. Kendrick-Jones. 1993. A myosin-like protein from a higher plant. J. Mol. Biol. 231: 148 154. 18 Molecular Motors in Plant Cells Kohno, T., S. Chaen, and T. Shimmen. 1990. Characterization of the translocator associated with pollen tube organelles. Protoplasma 154: 179 183. Kohno, T., R. Ishikawa, T. Nagai, K. Kohama, and T. Shimmen. 1992. Partial purification of myosin from lily pollen tubes by monitoring with an in vitro motility assay. Protoplasma 170: 77 85. Kohno, T., T. Okagaki, K. Kohama, and T. Shimmen. 1991. Pollen tube extract supports the movement of actin filaments in vitro. Protoplasma. 161: 75–77. Kohno, T., and T. Shimmen. 1987. Ca2 -induced F-actin fragmentation in pollen tubes. Protoplasma. 141: 177 179. Kohno, T. and T. Shimmen. 1988a. Accelerated sliding of pollen tube organelles along Characeae actin bundles regulated by Ca2. J Cell Biol. 106: 1539 1543. Kohno, T. and T. Shimmen. 1988b. Mechanism of Ca2 inhibition of cytoplasmic streaming in lily pollen tubes. J. Cell Sci. 91: 501 509. Krishnakumar, S. and D. G. Oppenheimer. 1999. Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development 126: 3079 3088. Lawrence, C. J., R. L. Malmberg, M. G. Muszynski, and R. K. Dawe. 2002. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54: 42 53. Lawrence, C. J., N. R. Morris, R. B. Meagher, and R. K. Dawe. 2001. Dyneins have run their course in plant lineage. Traffic 2: 362 363. Lazarowitz, S. G. and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535 548. Lee, Y.-R. J. and B. Liu. 2000. Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10: 797 800. Lee, Y. R., H. M. Giang, and B. Liu. 2001. A novel plant kinesin-related protein specifically associates with the phragmoplast organelles. Plant Cell 13: 2427 2439. Leinweber, B. D., P. C. Leavis, Z. Grabarek, C. L. Wang, and K. G. Morgan. 1999. Extracellular regulated kinase (ERK) interaction with actin and the calponin homology (CH) domain of actin-binding proteins. Biochem. J. 344: 117 123. Liu, B., R. J. Cyr, and B. A. Palevitz. 1996. A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell 8: 119 132. Liu, B. and Y. R. J. Lee. 2001. Kinesin-related proteins in plant cytokinesis. J. Plant Growth Regul. 20: 141 150. Liu, G.-Q., G. Cai, C. Del Casino, A. Tiezzi, and M. Cresti. 1994. Kinesin-related polypeptide is associated with vesicles from Corylus avellana pollen. Cell Motil. Cytoskel. 29: 155 166. Liu, L., J. Zhou, and T. C. Pesacreta. 2001. Maize myosins: diversity, localization, and function. Cell Motil. Cytoskeleton 48: 130 148. Lloyd, C., and P. Hussey. 2001. Microtubuleassociated proteins in plants–why we need a MAP. Nature Rev. Mol. Cell Biol. 2: 40 47. Lloyd, C. W. 1991. The Cytoskeletal Basis of Plant Growth and Form. New York: Academic Press, pp. 29 43. Long, R. M., R. H. Singer, X. Meng, I. Gonzalez, K. Nasmyth, and R. P. Jansen. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 277: 383 387. Lucas, W. J., S. Bouche-Pillon, D. P. Jackson, L. Nguyen, L. Baker, B. Ding, and S. Hake. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980 1983. Ma, Y.-Z. and L.-F. Yen. 1989. Actin and myosin in pea tendrils. Plant Physiol. 89: 586 589. Mas, P. and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol. 147: 945 958. Mas, P. and R. N. Beachy. 2000. Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc. Natl. Acad. Sci. USA 97: 12345 12349. Mascarenhas, J. P. 1990. Gene activity during pollen development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41: 317 338. Mascarenhas, J. P. 1993. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5: 1303 1314. Mathur, J., and N.-H. Chua. 2000. Microtubule stablization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12: 465 477. 465 466 References Mathur, J., N. Mathur, and M. Hulskamp. 2002. Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol. 128: 1031 1045. Mathur, J., P. Spielhofer, B. Kost, and N. Chua. 1999. The actin cytoskeleton is required to elaborate and maintain spatial patterning during trichome cell morphogenesis in Arabidopsis thaliana. Development 126: 5559 5568. Matsui, K., D. Collings, and T. Asada. 2001. Identification of a novel plant-specific kinesin-like protein that is highly expressed in interphase tobacco BY-2 cells. Protoplasma. 215: 105 115. Matthies, H. J. G., H. B. McDonald, L. S. B. Goldstein, and W. E. Theurkauf. 1996. Anastral meiotic spindle morphogenesis: role of the nonclaret disjunctional kinesin-like protein. J. Cell Biol. 134: 455 464. McLean, B. G., F. D. Hempel, and P. C. Zambryski. 1997. Plant intercellular communication via plasmodesmata. Plant Cell 9: 1043 1054. McLean, B. G., J. Zupan, and P. C. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101 2114. Mermall, V., P. L. Post, and M. S. Mooseker. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279: 527 533. Miller, D. D., S. P. Scordilis, and P. K. Hepler. 1995. Identification and localization of three classes of myosins in pollen tubes of Lilium longiflorum and Nicotiana alata. J. Cell Sci. 108: 2549 2563. Mitsui, H., S. Hasezawa, T. Nagata, and H. Takahashi. 1996. Cell cycle-dependent accumulation of a kinesin-like protein, KatB/C, in synchronized tobacco BY-2 cells. Plant Mol. Biol. 30: 177 181. Mitsui, H., K. Nakatani, K. Yamaguchi-Shinozaki, K. Shinozaki, K. Nishikawa, and H. Takahashi. 1994. Sequencing and characterization of the kinesin-related genes katB and katC of Arabidopsis thaliana. Plant Mol. Biol. 25: 865 876. Mitsui, H., K. Yamaguchi-Shinozaki, K. Shinozaki, K. Nishikawa, and H. Takahashi. 1993. Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of second- ary structure of KatA. Mol. Gen. Genet. 238: 362 368. Moepps, Y., S. Conrad, and H. Schraudolf. 1993. PCR-dependent amplification and sequence characterization of partial cDNAs encoding myosin-like proteins in Anemia phyllitidis (L.)Sw. and Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21: 1077 1083. Moscatelli, A., C. Del Casino, L. Lozzi, G. Cai, M. Scali, A. Tiezzi, and M. Cresti. 1995. High molecular weight polypeptides related to dynein heavy chains in Nicotiana tabacum pollen tubes. J. Cell Sci. 108: 1117 1125. Munchow, S., C. Sauter, and R. P. Jansen. 1999. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112: 1511 1518. Nagai, R. 1993. Regulation of intracellular movements in plant cells by environmental stimuli. Int. Rev. Cytol. 145: 251 310. Narasimhulu, S. B., Y.-L. Kao, and A. S. N. Reddy. 1997. Interaction of Arabidopsis kinesin-like calmodulin-binding protein with tubulin subunits: Modulation by Ca2 -calmodulin. Plant J. 12: 1139 1149. Narasimhulu, S. B. and A. S. N. Reddy. 1998. Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin-binding protein. Plant Cell 10: 957 965. Nebenfuhr, A., L. A. Gallagher, T. G. Dunahay, J. A. Frohlick, A. M. Mazurkiewicz, J. B. Meehl, and L. A. Staehelin. 1999. Stop-andGo movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121: 1127 1142. Nishihama, R., M. Ishikawa, S. Araki, T. Soyano, T. Asada, and Y. Machida. 2001. The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 15: 352 363. Nishihama, R. and Y. Machida. 2001. Expansion of the phragmoplast during plant cytokinesis: a MAPK pathway may MAP it out. Curr Opin Plant Biol. 4: 507 512. Nishihama, R., T. Soyano, M. Ishikawa, S. Araki, H. Tanaka, T. Asada, K. Irie, M. Ito, M. Terada, H. Banno, Y. Yamazaki, and Y. Machida. 2002. Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109: 87 99. 18 Molecular Motors in Plant Cells Oleynikov, Y. and R. H Singer. 1998. RNA localization: different zipcodes, same postman? Trends Cell Biol. 8: 381 383. Oppenheimer, D. G. 1998. Genetics of plant cell shape. Curr. Opin. Plant. Biol. 1: 520 524. Oppenheimer, D. G., M. A. Pollock, J. Vacik, D. B. Szymanski, B. Ericson, K. Feldmann, and D. Marks. 1997. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94: 6261 6266. Otegui, M. and L. A. Staehelin. 2000. Syncytialtype cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12: 933 947. Otegui, M. S., D. N. Mastronarde, B. H. Kang, S. Y. Bednarek, and L. A. Staehelin. 2001. Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13: 2033 2051. Parke, J., C. Miller, and B. H. Anderton. 1996. Higher plant myosin heavy-chain identified using a monoclonal antibody. Eur. J. Cell Biol. 41: 9 13. Perhonen, M., W. W. Sharp, and B. Russell. 1998. Microtubules are needed for dispersal of alpha-myosin heavy chain mRNA in rat neonatal cardiac myocytes. J. Mol. Cell Cardiol. 30: 713 722. Porter, J. A., B. Minke, and C. Montell. 1995. Calmodulin binding to Drosophila NinaC required for termination of phototransduction. EMBO J. 14: 4450 4459. Porter, J. A., M. Yu, S. K. Doberstein, T. D. Pollard, and C. Montell. 1993. Dependence of calmodulin localization in the retina on the NINAC unconventional myosin. Science 262: 1038 1042. Qiao, L., F. Grolig, P. P. Jablonsky, and R. E. Williamson. 1989. Myosin heavy chain: Detection by immunonblotting in higher plants and localization by immunofluorscence in the alga Chara. Cell Biol. Int. Rep. 13: 107 117. Radford, J. E. and R. G. White. 1998. Localization of a myosin-like protein to plasmodesmata. Plant J. 14: 743 750. Reddy, A. S. N. 2001a. Calcium: silver bullet in signaling. Plant Sci. 160: 381 404. Reddy, A. S. N. 2001b. Molecular motors and their functions plants. Intl. Rev. Cytol. & Cell Biol. 204: 98 179. Reddy, A. S. N. 2001c. Nuclear pre-mRNA processing in plants. CRC Cri. Rev. Plant Sci. 20: 523 572. Reddy, A. S. N. and I. S. Day. 2000. The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends Plant Sci. 5: 503 505. Reddy, A. S. N. and I. S. Day. 2001a. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2: 24.1 24.17. Reddy, A. S. N. and I. S. Day. 2001b. Kinesinlike proteins in Arabidopsis: a comparative analysis among eukaryotes. BMC Genomics 2: 2. Reddy, V. S. and A. S. N. Reddy. 1999. A plant calmodulin-binding motor is part kinesin and part myosin. Bioinformatics 10: 1055 1057. Reddy, A. S. N., S. B. Narasimhulu, F. Safadi, and M. Golovkin. 1996a. A plant kinesin heavy chain-like protein is a calmodulinbinding protein. Plant J. 10: 9 21. Reddy, A. S. N., F. Safadi, S. B. Narasimhulu, M. Golovkin, and X. Hu. 1996b. A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J. Biol. Chem. 271: 7052 7060. Reddy, V. S., F. Safadi, R. E. Zielinski, and A. S. N. Reddy. 1999. Interaction of a kinesinlike protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274: 31727 31733. Reichelt, S., A. E. Knight, T. P. Hodge, F. Baluska, J. Samaj, D. Volkmann, and J. Kendrick-Jones. 1999. Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 19: 555 567. Reilein, A. R., S. L. Rogers, M. C. Tume, and V. I. Gelfand. 2001. Regulation of molecular motor proteins. Intl. Rev. Cytol. Cell Biol. 204: 180 238. Rogers, G. C., C. L. Hart, K. P. Wedman, and J. M. Scholey. 1999. Identification of kinesinC, a calmodulin-binding carboxy-terminal kinesin in animal (Strongylocentrotus purpuratus) cells. J. Mol. Biol. 294: 1 8. Samuels, A. L., J. T. H. Giddings, and L. A. Staehelin. 1995. Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130: 1345 1357. 467 468 References Sawin, K. E. and T. J. Mitchison. 1995. Mutations in the kinesin-like protein Eg5 disrupting localization to the spindle. Proc. Natl. Acad. Sci. USA 92: 4289 4293. Shimmen, T. and E. Yokota. 1994. Physiological and biochemical aspects of cytoplasmic streaming. Int. Rev. Cytol. 155: 97 140. Siddiqui, S. S. 2002. Metazoan Motor Models: Kinesin superfamily in C. elegans. Traffic 3: 20 28. Smirnova, E., A. S. N. Reddy, J. Bowser, and A. S. Bajer. 1998. A minus end-directed kinesin-like motor protein, KCBP, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil. Cytoskeleton. 41: 271 280. Smirnova, E. A. and A. S. Bajer. 1992. Spindle poles in higher plant mitosis. Cell Motil. Cytoskelet. 23: 1 7. Song, H., M. Golovkin, A. S. N. Reddy, and S. A. Endow. 1997. In vitro motility of AtKCBP, a calmodulin-binding kinesin-like protein of Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 322 327. Staehelin, L. A. and P. K. Hepler. 1996. Cytokinesis in higher plants. Cell 84: 821 824. Staiger, C. J. and M. Schilwa. 1987. Actin localization and function in higher plants. Protoplasma 141: 1 12. Strompen, G., F. El Kasmi, S. Richter, W. Lukowitz, F. F. Assaad, G. Jurgens, and U. Mayer. 2002. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycledependent manner. Curr. Biol. 12: 153 158. Sylvester, A. W. 2000. Division decisions and the spatial regulation of cytokinesis. Curr. Opin. Plant Biol. 3: 58 66. Szymanski, D. B., D. M. Marks, and S. M. Wick. 1999. Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell. 11: 2331 2348. Takagi, S. and R. Nagai. 1986. Intracellular Ca2 concentration and cytoplasmic streamig in Vallisneria mesophyll cells. Plant Cell Physiol. 27: 953. Takizawa, P. A., A. Sil, J. R. Swedlow, I. Herskowitz, and R. D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389: 90 93. Tamura, K., K. Nakatani, H. Mitsui, Y. Ohashi, and H. Takahashi. 1999. Characterization of katD, a kinesin-like protein gene specifically expressed in floral tissues of Arabidopsis thaliana. Gene 230: 23 32. Tang, X. J., P. K. Hepler, and S. P. Scordilis. 1989. Immunochemical and immunocytochemical identification of a myosin heavy chain polypeptide in Nicotiana pollen tubes. J. Cell Sci. 92: 569 574. Tiezzi, A., A. Moscatelli, G. Cai, A. Bartalesi, and M. Cresti. 1992. An immunoreactive homolog of mammalian kinesin in Nicotiana tabacum pollen tubes. Cell Motil. Cytoskel. 21: 132 137. Tominaga, Y., T. Shimmen, and M. Tazawa. 1983. Control of cytoplasmic streaming by extracellular Ca2 in permeabilized Nitella cells. Protoplasma 116: 75 77. Vahey, M., M. Titus, R. Trautwein, and s. Scordilis. 1982. Tomato actin and myosin: Contractile proteins from a higher land plant. Cell Motil. 2: 131 148. Van Gestel, K., R. H. Kohler, and J. P. Verbelen. 2002. Plant mitochondria move on F-actin, but their positioning in the cortical cytoplasm depends on both F-actin and microtubules. J. Exp. Bot. 53: 659 667. Vidali, L. and P. K. Hepler. 2001. Actin and pollen tube growth. Protoplasma 215: 64 76. Vos, J. W., F. Safadi, A. S. Reddy, and P. K. Hepler. 2000. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12: 979 990. Wang, D. Y., S. Kumar, and S. B. Hedges. 1999. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. R. Soc. Lond. B. Biol. Sci. 266: 163 171. Wang, W., D. Takezawa, S. B. Narasimhulu, A. S. N. Reddy, and B. W. Poovaiah. 1996. A novel kinesin-like protein with a calmodulinbinding domain. Plant Mol. Biol. 31: 87 100. White, R. G., K. Badelt, R. L. Overall, and M. Vesk. 1994. Actin associated with plasmodesmata. Protoplasma. 180: 169 184. Wick, S. M. 1991. Spatial aspects of cytokinesis in plant cells. Curr. Opin. Cell. Biol. 3: 253 260. Williamson, R. E. 1976. Actin and Motility in Plant Cells. Amsterdam: North-Holland Publ., pp. 91 101. Williamson, R. E. 1993. Organelle movements. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44: 181 202. 18 Molecular Motors in Plant Cells Williamson, R. E. and C. C. Ashley. 1982. Free Ca2 and cytoplasmic streaming in the alga Chara. Nature 296: 647 650. Woehlke, G. and M. Schliwa. 2000. Walking on two heads: the many talents of kinesin. Nature Reviews: Mol. Cell Biol. 1: 50 58. Wolenski, J. S. 1995. Regulation of calmodulinbinding myosins. Trends Cell Biol. 5: 310 316. Yamamoto, K., S. Hamada, and T. Kashiyama. 1999. Myosins from plants. Cell. Mol. Life Sci. 56: 227 232. Yamamoto, K., M. Kikuyama, N. Sutoh-Yamamoto, and E. Kamitsubo. 1994. Purification of actin based motor protein from Chara corallina. Proc. Japan Acad. 70: 175 180. Yamamoto, K., M. Kikuyama, N. Sutoh-Yamamoto, E. Kamitsubo, and E. Katayama. 1995. Myosin from alga Chara: Unique structure revealed by electron microscopy. J. Mol. Biol. 254: 109 112. Yamashita, R. A., J. R. Sellers, and J. B. Anderson. 2000. Identification and analysis of the myosin superfamily in Drosophila: a database approach. J. Muscle Res. Cell Motil. 21: 491 505. Yasuhara, H., S. Snobe, and H. Shibaoka. 1993. Effects of taxol on the development of cell plate and the phragmoplast in tobacco BY-2 cells. Plant Cell Physiol. 34: 21 29. Yokota, E., A. R. McDonald, B. Liu, T. Shimmen, and B. A. Palevitz. 1995. Localization of a 170 kDa myosin heavy chain in plant cells. Protoplasma 185: 178 187. Yokota, E. and T. Shimmen. 1994. Isolation and characterization of plant myosin from pollen tubes of lily. Protoplasma 177: 153 162. Yokota, E. and T. Shimmen. 1999. The 135-kDa actin-bundling protein from lily pollen tubes arranges F- actin into bundles with uniform polarity. Planta 209: 264 266. Yokota, E., S. Muto, and T. Shimmen. 1999a. Inhibitory regulation of higher-plant myosin by Ca2 ions. Plant Physiol. 119: 231 240. Yokota, E., C. Yukawa, S. Muto, S. Sonobe, and T. Shimmen. 1999b. Biochemical and immunocytochemical characterization of two types of myosins in cultured tobacco bright yellow-2 cells. Plant Physiol. 121: 525 534. 469