* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Genetics(Semester(One,(Year(Two!
Frameshift mutation wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Public health genomics wikipedia , lookup
Minimal genome wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genetic engineering wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Human genetic variation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Y chromosome wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Genetic drift wikipedia , lookup
Genomic imprinting wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Neocentromere wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Population genetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression programming wikipedia , lookup
Helitron (biology) wikipedia , lookup
Genome editing wikipedia , lookup
Designer baby wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Genome (book) wikipedia , lookup
X-inactivation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
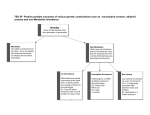
Genetics(Semester(One,(Year(Two! Need$genetic$variation$ • • Natural$ o Monogenic$ o Polygenic$ Induced$ o Mutagenesis$ Variation$between$organisms$ 1. Phenotypic$ a. Morphological$ b. Sterile$ c. Lethal$ 2. Molecular$ a. Biochemical$(enzymes,$hormones$etc)$ b. DNA$ c. Proteins$ Types&of&alleles& 1. 2. 3. 4. 5. 6. Wild$type$–$dominant$phenotype$(normal)$ Hypomorphic$mutant$–$defective$phenotype,$still$partially$effective$ Amorphic$mutant$–$loss$of$function,$extreme$change$ Hypermorphic$mutant$–$increase$of$function,$more$efficient$ Neomorphic$mutant$–$gain$of$function,$new/different$function$ Antimorphic$mutant$(dominant$negative$mutation)$–$altered$function,$antagonistic$to$wild$type.$Works$ against$wild$type$protein.$ $ 7. Conditional$mutant$–$different$under$certain$conditions$e.g.$temperature$sensitive$mutant$ Penetrance& Mutant$allele$in$genetic$background,$depending$on$the$number$of$mutants$or$how$many$genes$are$interacting$the$ presence$of$mutations$may$have$varied$effects$ • • Example$One$ o Wild$type$–$B,$C$ o Mutant$–$A$ o Slightly$abnormal$phenotype$ $Example$Two$ o Wild$type$–$B$ o Mutant$–$A,$C$ o Significantly$abnormal$phenotype$ Mutant$alleles$do$not$have$to$express$phenotype,$can$be$affected$by$gene$interaction$(penetrance,$degree$of$ expression)$ $ $ Dominance& Only$one$copy$of$allele$needed$to$produce$phenotype$ Dominant$allele$phenotype$may$have$partial$or$total$gene$product$from$dominant$allele$ E.g.$AA:$100%$dominant,$Aa:$50%$dominant$50%$recessive.$But$both$express$dominant$phenotype$ Haploinsufficiency:$May$need$100%$gene$product$to$express$phenotype.$If$heterozygous,$not$enough$gene$product$ of$recessive$allele$so$expresses$dominant$phenotype.$ A$hypermorphic/neomorphic$allele$may$lead$to$homozygous$extreme$mutant$compared$to$the$heterozygous.$ Amorphic$mutation:$Loss$of$function,$usually$recessive$ Antimorphic$mutation/Neomorphic$mutation:$Usually$dominant$ Codominance:$Heterozygous$expresses$phenotype$of$both$homozygotes$e.g$blood$antigens$ Incomplete$Dominance:$Heterozygous$has$intermediate$phenotype$e.g.$flower$colours$$$ Mutant$alleles$may$show$a$dominant$phenotype$when$heterozygous$but$a$different$(recessive)$phenotype$when$ homozygous.$This$is$due$to$gene$product$interactions.$Heterozygous$means$two$different$gene$products$may$affect$ each$other,$homozygous$is$only$one$gene$product$(may$give$different$phenotype)$ At$the$DNA$sequence$level$all$alleles$are$co\dominant.$At$a$molecular$level,$protein$production$sequence.$ Variations$in$the$DNA$sequence$produce$equal$amounts$of$protein,$no$dominance.$$ Whether$an$allele$shows$dominance$or$not$depends$on$which$aspect$of$the$phenotype$you$are$studying.$ The$phenotype$(morphological$variation)$not$the$allele$is$dominant$or$recessive.$ $ Mendel’s&Laws& 1. Alleles$will$always$segregate$away$from$each$other$into$gametes.$Aa$allele$to$A$+$a$gamete$contribution$ 2. Alleles$of$separate$genes$will$always$segregate$independently$into$gametes.$ $ $ !! !! !! !! × × !! !! !! !! = 9: 3: 3: 1!!ℎ!"#$%&!!!"#$%.$ = 1: 1: 1: 1!!ℎ!"#$%&!!!"#$%$ Chromosome&Theory& • • • • #$of$phenotypes$is$exponential,$#$of$heritable$factors$exceeds$the$#$of$chromosomes$ Therefore$expect$multiple$factors$to$exist$per$chromosome$ Predict$that$factors$on$the$same$chromosome$will$be$transmitted$as$a$block$ Therefore$factors$on$the$same$chromosome$will$not$assort$independently$ Test&cross&of&Drosophila& Recombinant:$new$genotype$ Combinant:$parental$genotype$ • • • • Two$traits,$red/purple$eyes$and$normal/vestigial$wings$ Parents:$Homozygous$wild$type$x$homozygous$recessive$ F1:$Heterozygous$wild$type$ F1$crossed$with$homozygous$recessive$(test$cross)$ o • • • !" ! !"! !" !" × !" !" !" !" $$ o F1$produces$>1$type$of$gamete,$tester$produces$only$1$type$of$gamete$ Mendel$would$predict$equal$proportions$of$F1$gametes$(!" ! !"! , !"!!", !" ! !", !"!"! )!$due$to$ independent$assortment,$thus$giving$equal$proportion$of$phenotypes$ Chromosome$theory$would$predict$equal$proportions$if$gene$loci$were$on$different$chromosomes$or$only$ combinants$if$they$were$on$the$same$chromosome$ Observed$presence$of$all$four$phenotypes$with$ratio’s$heavily$stacked$to$parental$phenotypes.$$ (Phenotypes$not$genotypes)$ Ab$and$aB$would$be$parentals$ AB$and$ab$would$be$recombinants$ BUT$we$only$know$that$the$ab$phenotypes$are$recombinants$for$sure$(AB$phenotypes$may$have$aB,$Ab,$or$AB$ gametes)$ The$frequency$of$the$ab$phenotypes$is$the$probability$of$the$ab$gametes$squared.$(Pr !"× Pr !")$ Therefore$frequency$of$ab$gametes$is$square$root$of$0.01$=$0.1$ Frequency$of$the$other$possible$recombinant$AB$is$equal$in$frequency$as$when$making$ab,$AB$is$made$as$well.$$ Recombination$frequency$= (!.!!!.!) !.! = 0.2 = 20% = 20!!"!(!"#!!"#$%)$ $ Physical&Linkage&vs&Genetic&Linkage& Genes$are$linked$if$0%<RF<50%$ 0% ≤ !"#$%&'()*'$(!!"#$%#&'( ≤ 50%$ If$genes$are$far$apart$on$the$same$chromosome,$they$might$independently$assort$ To$show$linkage$for$genes$located$far$apart$(RF$~$50%)$we$can$$ 1. Take$advantage$of$achiasmatic$gametogenesis$ 2. Find$a$third$gene$in$the$middle$ Frequencies$of$crossing$over$is$proportional$to$distance$between$genes$ Can$combine$data$from$different$experiments$to$form$a$chromosome$map$ $ $ $ $ Can$be$re\organised$into$ $ $ $ $ Can$also$build$a$chromosome$map$by$combining$multiple$two$factor$crosses.$But$less$accurate$because$may$miss$$ double$recombinations$$ Look$at$the$largest$map$distance,$the$two$genes$involved$will$be$the$first$and$last$in$the$gene$order$ $ $ Genome&maps& Allow$the$tracking$of$chromosome$segments.$$ Know$where$to$look$for$certain$defects$ Limitations#of#phenotypic#markers# 1. 2. 3. 4. Phenotypic$mutants$are$rare$ Differences$are$not$always$clear$cut$ Multiple$genes$may$affect$one$phenotypes$ The$need$to$maintain$mutant$stocks$is$expensive$ Molecular#markers# • • • DNA$ o Differences$in$DNA$sequence$ o Differences$in$charge$of$proteins$ Both$phenotypic$and$molecular$tag$and$track$chromosome$transmission$ Scored$by$chemical$assays$$ Benefits#of#Molecular#markers# 1. Abundant$ 2. Random$distribution$ a. Provides$genome$wide$coverage$ 3. Many$markers$can$be$scored$(genotyped)$in$a$single$cross$ 4. Genotyping$can$be$automated$and$streamlined$ Markers:$Polymorphism$+$Detection$method$ Polymorphisms$ • • • • Nucleotide$substitution$ o Single$nucleotide$polymorphism$(SNP)$ Insertion/deletion$ Expansion/contraction$ Microsatellites$$ o Base$sequence$repetitions$$ Detection$methods$ Phage&Genetics& • • • • Phage$are$haploid$$ o Phenotype$will$equal$genotype$ No$nuclei,$do$not$undergo$mitosis$or$meiosis$ Undergo$recombination$and$complementation$ Have$genetic$variation$ Phage#Genetic#Variation# 1. Unconditional$Mutants$ a. Plaque$morphology$mutants$ i. Clear$plaque$mutants$ ii. Lysogen$negative$mutants$ 2. Conditional$Mutants$ a. Grow$in$one$set$of$conditions$known$as$the$permissive$conditions$ b. Will$not$grow$under$non\permissive/restrictive$conditions$ c. Temperature$sensitive$mutants$ d. Host$range$specific$mutants$ Plaque#morphology#mutants# • • • λ$phage$that$have$lost$the$ability$to$form$lysogens$(always$undergo$lytic$cycle)$ $Show$plaques$that$do$not$have$any$lysogens$growing$in$them.$Always$clear$ Rapid$lysis$(r_$)mutants$of$phage$T4$ Temperature#sensitive#mutants# • E.g.$grow$at$37$degrees$but$not$at$42$degrees$ Host#range#mutants# • E.g$Unable$to$grow$on$certain$strains$of$E.Coli$ $ Recombination&in&phage$ 1. Co\infection$of$host$cell$with$multiple$phages.$ a. Single$bacteria$infected$with$multiple$mutations$ 2. Co\infection$creates$‘diploid$state’$for$the$phage$ 3. Recombination$event$occurs$between$two$phage$DNA$molecules$with$different$mutations$ 4. Two$distinct$phage$DNA$molecules$are$created,$different$from$the$parentals$ 5. The$progeny$will$have$different$genotypes$than$the$parents$ $