* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Hsp90
Signal transduction wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Biochemistry wikipedia , lookup
Paracrine signalling wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Metalloprotein wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Expression vector wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Interactome wikipedia , lookup
Western blot wikipedia , lookup
Protein purification wikipedia , lookup
Two-hybrid screening wikipedia , lookup
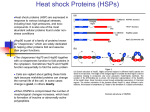
Protein Folding and Molecular Chaperones Contents Protein Folding Molecular Chaperones Protein Folding and Diseases 1. Protein Folding Protein-Folding Problem 1958 John Kendrew et al., published the first structure of a globular protein, myoglobin. “ Perhaps the most remarkable features of the molecule are its complexity and its lack of symmetry” 1962 Nobel prize in Chemistry was awarded to Max Perutz and John Kendrew. Now ~80,000 structures in protein database (PDB) Common Structural Patterns Motifs, folds, or supersecondary structures Stable arrangements of several elements of secondary structure Domains Stable, globular units Classification of Protein Structures Structural classification of proteins (SCOP) database Classification All a All b a/b : a and b segments are interspersed or alternate a + b : a and b regions are segregated ~1,200 different folds or motifs Protein family (~4,000) Proteins with similarities in Primary sequence (and/or) Structure Function Superfamily Families with little primary sequence similarity but with similarities in motifs and function Structural classification from SCOP database Structural classification from SCOP database Amino Acid Sequenc Determines Tertiary Structure Amino acid sequence contains all the information required to protein folding First experimental evidence by Christian Anfinsen (1950s) Denaturation of ribonuclease with urea and reducing agent Spontaneous refolding to an active form upon removal of the denaturing reagents Protein Folding is not a trialand-error process E. coli make 100 a.a. protein in 5 sec 10 possible conformations/ a.a. 10100 conformations 10-13 sec for each conformation 1077 years to test all the conformations Protein folding problem has not yet been solved The physical folding code How is the 3D structure determined by the physicochemical properties encoded in the amino acid sequence? The folding mechanism How can proteins fold so fast even with so many possible conformations? Predicting protein structure using computers Can we devise a computer algorithm to predict 3D structure from the amino acid sequnece? The Physical Code of Protein Folding Weak Interactions Hydrogen bond Hydrophobic interactions Van der Waals interactions Electrostatic interactions Backbone angle preferences Chain entropy Large loss of chain entropy upon folding Covalent bonding Disulfide bonding The Rate Mechanisms of Protein Folding Models for protein folding Hierarchical folding From local folding (a helix, b sheets) to entire protein folding Molten globule state model Initiation of folding by spontaneous collapse by hydrophobic interactions Thermodynamics of Protein Folding Free-energy funnel Unfolded states High entropy and high free energy Folding process Decrease in the number of conformational species (entropy) and free energy Semistable folding intermediates Computing Protein Structures Computer-based protein-structure prediction competition Critical Assessment of protein Structure Prediction (CASP) in every second summer since 1994 Computing Protein Structures Template-modeling (homology modeling, comparative modeling) Structure prediction based on the structure of a protein with a sequence homology. Free modeling (ab initio, de novo modeling) Fragment assembly PDB search of overlapping fragments of target proteins Assembly of fragments using some scoring functions Successful for short proteins (<100 a.a.) Computing Protein Structures 2. Molecular Chaperones Molecular Chaperones Proteins facilitating protein folding, transport, and degradation AAAA Misfolding Folding Translocation Folding Degradation Nuclear import/export proteasome Classes of Molecular Chaperones Ribosome-associated chaperones Trigger factor (prokaryotes) RAC, NAC (eukaryotes) Cytosolic chaperones Hsp70 Induced in stressed cells (heat shock protein) Binding to hydrophobic regions of unfolded proteins, preventing aggregation Cyclic binding and release of proteins by ATP hydrolysis and cooperation with co-chaperones (Hsp40 etc.) E. coli: DnaK (Hsp70), DnaJ (Hsp40) Hsp90 Small Hsps Chaperonin Protein complex providing microenvironments for protein folding E. coli : 10~15% protein require GroES (lid) and GroEL Eukaryotes: TriC/CCT Organelle-specific chaperones (eukaryotes only) ER chaperones Mitochondrial chaperones Isomerases in Protein Folding Protein disulfide isomerase (PDI) Shuffling disulfide bonds Peptide prolyl cis-trans isomerase (PPI) Interconversion of the cis and trans isomers of Pro peptide bonds Co-translational Folding Prokaryotes Trigger factor Cyclic association and dissociation with ribosome Binds to hydrophobic regions of newly synthesized polypeptide chains Shields nascent chains from degradation by proteases Improve the yields of correctly folded model substrates by reducing the speed of folding Eukaryotes Hsp70 and J-protein–based systems Ribosome-associated complex (RAC) Heterodimeric nascent polypeptide-associated complex (NAC). Trigger Factor Molecular Chaperones Protein Folding by DnaK and DnaJ Chaperonin in Protein Folding Roles of Hsp70 and Hsp90 AAAA Misfolding Folding Hsp70 Metastable client proteins Hsp90 Assembly Hsp70 Hsp90 Folding Hsp90 Prokaryotes HtpG Yeast Hsp82, Hsc82 Higher eukaryotes HtpG, Trap-1 N Other Hsp90s N Hsp90a, Hsp90b (cytosol) Grp94 (ER) Trap-1 (mitochondria) M C M Acidic linker C Hsp90 Chaperone Network in Yeast Genes interacting with Hsp90 ~200 physical interactions ~451 genetic and chemical-genetic interactions Zhao, R. et al., 2005, Cell McClellan A.J. et al., 2007, Cell Hsp90 Chaperone Cycle Open ATP-bound Lid N ATP M C Lid open : ADP bound ADP + Pi ATP hydrolysis Closed Lid close : ATP bound Class of Hsp90 Co-chaperones With TPR Without TPR Higher eukaryotes Yeast Function Hop Sti1 Adaptor to Hsp70 Cyp40 Cpr6, Cpr7 Peptidyl-prolyl isomerase FKBP51, FKBP52 - Peptidyl-prolyl isomerase Sgt1 Sgt1 Adaptor for SCF and client proteins PP5 Ppt1 Phosphatase Aha1 Aha1, Hch1 Activation of Hsp90 ATPase activity p23 Sba1 Inhibition of Hsp90 ATPase activity Cdc37 Cdc37 Adaptor for kinases Chp-1, Melusin - Unknown TPR Motif Protein-protein interaction module One TPR motif contains two antiparallel a-helices Tandem array of TPR motifs generate a righthanded helical structure TPR domain in co-chaperones binds to MEEVD sequence in the Hsp90 C-terminus Hsp90 MEEVD PP5 TPR Regulation of ATPase Activity by Co-chaperones Sgt1 (CS domain) Hsp90 N (O) ATP ADP + Pi Hsp90 N (O) Cdc37 (125-378) Hsp90 N (C) Sba1 Aha1 (1-153) Protein Folding and Aggregation Conditions Inducing Protein Aggregation Mutations prone to aggregate Huntington’s disease Familial forms of Parkinson’s disease and Alzheimer’s disease Defects in protein biogenesis Translational errors Assembly defects of protein complexes Environmental stress conditions Heat shock Oxidative stress Aging Deposition of Aggregates Bacteria Inclusion body Yeast Juxtanuclear quality-control compartment (JUNQ) Soluble, misfolded, ubiquitylated proteins Perivacuolar insoluble protein deposit (IPOD). Insoluble, terminally aggregated Mammals Aggresome Protein Disaggregation Hsp70-Hsp104 (ClpB) bi-chaperone Hsp70-J protein Transfer aggregates to Hsp104 Hsp104 Threading activity to refold aggregate 3. Protein Folding and Diseases Protein-Folding Diseases Amyloidoses Diseases caused by formation of insoluble amyloid fibers Protein-Folding Diseases Cystic fibrosis Misfolding of cystic fibrosis transmembrane conductance regulator (CFTR) Neurodegenerative diseases Alzheimer’s, Parkinson’s, Huntinton’s disease, ALS Prion diseases Mad cow disease (bovine spongiform encephalopathy, BSE) Kuru, Creutsfeldt-Jakob disease in human Scrapie in sheep Prion : proteinaceous infectious only protein PrPSc (scrapie) prion form converts PrPC to PrPSc Formation of Amyloid Fibers Amyloid –b peptide Chaperones as Drug Targets Hsf1 Transcriptional activation of heat shock proteins Activators of Hsf1 as drugs for proteinfolding diseases Hsp70 Hsp90 Clients proteins include some oncoproteins Hsp90 inhibitors as cancer drugs Hsp90 Client Proteins in Cancer Hsp90 client protein Roles of Hsp90 client proteins in cancer Her2, Raf-1, Akt Self-sufficiency in growth signals Plk, Wee1, Myt1 Insensitivity to antigrowth signals RIP, Akt Evasion of apoptosis hTERT Limitless replicative potential Hif-1a, Fak, Akt Sustained angiogenesis Met Tissue invasion and metastasis Hsp90 Inhibitors as Anti-Cancer Drug Her2,Raf-1,Akt, Hif-1a, survivin, mutant p53 Oncoprotein Protein stabilization Oncoprotein Cancer Hsp90 inhibitor Bended Form of ATP & 17-DMAG in the Pocket J.M. Jez et al, 2003, Chemistry & Biology Hsp90 Inhibitors Geldanamycin ATP Radicicol PU3 17-AAG