* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download How to obtain a clone of a specific gene
Gel electrophoresis of nucleic acids wikipedia , lookup
Non-coding DNA wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene expression wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
Molecular cloning wikipedia , lookup
Gene nomenclature wikipedia , lookup
List of types of proteins wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Promoter (genetics) wikipedia , lookup
SNP genotyping wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Gene regulatory network wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Molecular evolution wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Genomic library wikipedia , lookup
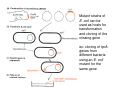
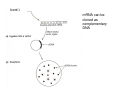
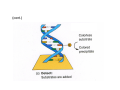
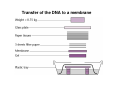
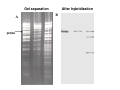
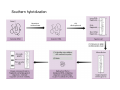
How to obtain a clone of a specific gene There are two basic strategies: -Direct selection for the desired clone - Identification of the clone from a gene library (several techniques can be employed) Direct selection for the kanamycin resistance gene Mutant strains of E. coli can be used as hosts for transformation and cloning of the missing gene ex: cloning of tprA genes from different bacteria using an E. coli mutant for the same gene The scope and limitations of marker rescue Limitations -a mutant strain must be available for the gene in question -a medium on which only the wild-type can survive is needed Applications -most genes that code for biosynthetic enzymes -is not restricted to E. coli since many auxotrophs from other bacteria and yeast are available Identification of a clone from a gene library: preparation of a bacterial gene library In eukaryotes different genes are expressed in different types of cell Preparation of a cDNA gene library from eukaryotes (cont.) mRNA can be cloned as complementary DNA Identification of a clone from a gene library: Method: nucleic acid hybridization Any two singlestranded nucleic acid molecules have the potential to form base pairs with one another Hybridization mechanism (cont.) Southern hybridization (DNA-DNA hybridization) E.M.Southern, 1970 Transfer of the DNA to a membrane (cont.) After hybridization Gel separation A probe B Southern hybridization Colony hybridization (developed in the late 1970) (cont.) Colony hybridization -The colonies are transferred to a nitrocellulose or nylon membrane -The membrane/immobilized cells is treated to remove all contaminating material, leaving just DNA denatured -The DNA is bound to the membrane by heat or UV light -The labelled probe is denatured and hybridization taskes place -Washing steps to remove unbound probe -Probe detection by autoradiography, colorimetry or chemoluminescence Probes -Can be labelled with radioactive nucleotides or with non-radioactive (biotin, digoxigenin, fluorescein, etc) -The probes have to share at least a part of the sequence of the molecule of interest to detect -The size of the probes is usually between 100-1000 bp Labelling DNA by random priming The mixture of hexameric oligonucleotides of random sequence is sufficiently complex to include at least a few molecules that can base pair with the target sequence so that the polymerases synthesize the complementary strand with at least one labelled dNTP Non-radioactive labelling of DNA probes Examples of the practical use of a hybridization probe Heterologous probing makes use of hybridization between related sequences for clone identification Identification of pgmG from S. elodea Box I Active site PgmG S. paucimobilis (AF167367) ExoC Azospirillum brasilense (P45632) PmmA Prochlorothrix hollandica (AAC44309) ExoC Rickettsia prowazekii (CAA14961) AlgC Pseudomonas aeruginosa (P26276) Pgm Neisseria gonorrhoeae (P40390) XanA Xanthomonas campestris (AAA27610) ManB Escherichia coli (P24175) RFK7 E. coli (P37742) CelB A.xylinum (P38569) Pgm E. coli (P36938) Pgm1 Homo sapiens (P36871) Pgm1 Saccharomyces cerevisiae (S41199) PgmU Oryctolagus cuniculus (P00949) Box II Box III Metal-ion-binding Sugar binding QITGSHNPGN-109 MITGSHNPPD-111 QITGSHNPPE-110 MVTGSHNPRD-116 MLTGSHNPPD-113 MITGSHNPPD-108 MVTASHNPMD-102 EVTASHNPMD-103 EVTASHNPMD-101 GADGDGDRID-249 GFDGDGDRIG-247 AFDGDGDRIG-251 AFDGDGDRIG-296 AFDGDGDRVG-249 AFDGDADRLG-246 AWDGDFDRCF-244 AFDGDFDRCF-252 AFDGDFDRCF-250 LAGEMSGH-326 LAGEMSGH-327 LAGEMSGH-331 LAGEMSGH-376 LAGEMSGH-329 VAGEMSGH-326 YGGEMSAH-324 YGGEMSAH-332 YGGEMSAH-329 VITPSHNPPE-153 VITPSHNPPE-151 ILTASHNPGG-122 ILTASHNPGG-125 ILTASHNPGG-122 ANDTDADRHG-313 ANDPDYDRHG-311 AFDGDGDRNM-295 ASDGDGDRNM-298 AFDGDGDRNM-295 FGGEESAG-397 FGGEESAG-394 LCGEESFG-380 ICGEESFG-383 LCGEESFG-380 * **** * * ** ** * Degenerated primers based on known proteins with phosphoglucomutase activity to amplify an internal fragment of pgmG followed by screening of the Sphingomonas elodea genomic library