* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download How hereditary information is stored in the genome.
Human genetic variation wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene expression programming wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene therapy wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
Gene nomenclature wikipedia , lookup
Non-coding DNA wikipedia , lookup
Minimal genome wikipedia , lookup
Gene desert wikipedia , lookup
Whole genome sequencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Metagenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Genome (book) wikipedia , lookup
Public health genomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Human genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
Human Genome Project wikipedia , lookup
Genomic library wikipedia , lookup
Microevolution wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Helitron (biology) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
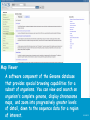
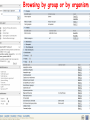
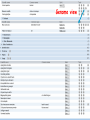
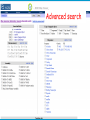
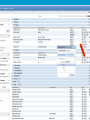
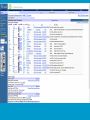
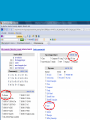
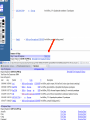
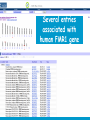
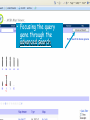
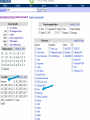
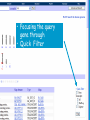
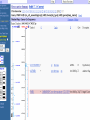
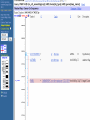
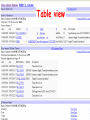
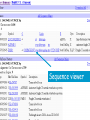
How hereditary information is stored in the genome. Three types of maps : – Linkage maps of genes – Banding pattern of chromosome – DNA sequences These maps lie on three very different types of data: – Observed pattern of heredity; gene or lingake group transmission and recombination frequency. – Identification of physical bands – Physical sequence of nucleotides High resolution maps directly based on DNA markers. A marker is any sequence variation among individual that exists at a defined position within the genome and can be associated with individual phenotypic characteristics. The very great achievment of biology is to find connections between markers and phenotypes. Map Viewer Software to visualize genomic or chromosomic maps Map Viewer A software component of the Genome database that provides special browsing capabilities for a subset of organisms. You can view and search an organism's complete genome, display chromosome maps, and zoom into progressively greater levels of detail, down to the sequence data for a region of interest. LINK RETE Map Viewer Utility 1. To display maps from different types of data; 2. To compare different kinds of data; 3. To follow links between them; 4. To compare map information from different organisms. Map Viewer access 1. Browsing by organism 2. Following links from various data base (Nucletide, Gene, UniSTS..) Browsing by group or by organism Genome view Advanced search LOCALIZE THE CFTR GENE IN THE MOUSE GENOME Using MapViewer to localize human FMR1 gene • From the Map Viewer homepage, select Homo sapiens (human), enter FMR1. • On the resulting genome page several entries are returned. Several entries associated with human FMR1 gene • Focusing the query gene through the advanced search 3 hits • Focusing the query gene through • Quick Filter Table view Sequence viewer Genomic contig Map selection through maps and options How can I obtain the genomic sequence around my gene of interest? 1. 2. 3. 4. Enter in MapViewer home page. Refine search through advanced search. Select the map (gene). Dowload/View Sequence/Evidence Map Viewer access 1. Browsing by organism 2. Following links from various data base Map Viewer access Following links from various data base OMIM (External Links Genome) Nucleotide (Related information) Gene (Links) UniSTS (in Mapping Information)