* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download regulatory-network
Biology and consumer behaviour wikipedia , lookup
Gene desert wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Point mutation wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
History of genetic engineering wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Microevolution wikipedia , lookup
Minimal genome wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome (book) wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome evolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
Designer baby wikipedia , lookup
Gene expression profiling wikipedia , lookup
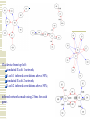
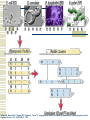
Gene Regulatory Network Inference Progress in Disease Treatment Personalized medicine is becoming more prevalent for several kinds of cancer treatment 10-Feb-2009 – Breast Bioclassifier developed at the Huntsman Cancer Institute 1/8 women will be diagnosed with breast cancer Microarray analysis can separate large group who need no treatment Savings in cost and lifestyle With $100 human genomes, doctors can determine which drugs will be effective for your genotype Biological Networks Gene regulatory network: two genes are connected if the expression of one gene modulates expression of another one by either activation or inhibition Protein interaction network: proteins that are connected in physical interactions or metabolic and signaling pathways of the cell; Metabolic network: metabolic products and substrates that participate in one reaction; Background Knowledge Cell reproduction, metabolism, and responses to the environment are all controlled by proteins; Each gene is responsible for constructing a single protein; Some genes manufacture proteins which control the rate at which other genes manufacture proteins (either promoting or suppressing); Hence some genes regulate other genes (via the proteins they create) ; What is Gene Regulatory Network? Gene regulatory networks (GRNs) are the on-off switches of a cell operating at the gene level. Two genes are connected if the expression of one gene modulates expression of another one by either activation or inhibition An example. Sources: http://www.ornl.gov/sci/techresources/Human_Genome/graphics/slides/images/REGNET.jpg Why Study GRN? Genes are not independent; They regulate each other and act collectively; This collective behavior can be observed using microarray; Some genes control the response of the cell to changes in the environment by regulating other genes; Potential discovery of triggering mechanism and treatments for disease; Learning Causal Relationships High-throughput genetic technologies empowers to study how genes interact with each other; If gene A consistently turns on after Gene C, then gene C may be causing gene A to turn on We have to have a lot of carefully controlled time series data to infer this Kegg http://www.genome.jp/kegg/pathway.html Pathgen Microarray data Genes Samples Gene up-regulate, down-regulate; Learning from microarray data Recurrent Neural Networks Bayesian learning approaches AIRnet: Asynchronous Inference of Regulatory networks 1. Classify gene levels using k-means clustering 2. Compute influence vectors (i.v.) 3. Convert i.v.'s into a sorted list of edges 4. Use Kruskal's algorithm to find the minimum-cost spanning tree Influence Vectors 1. Perform pairwisecomparisons of change in gene levels between samples, adding or subtracting from i.v. 2. Divide i.v. by the total number of comparisons Clockwise from top left: simulated E.coli 1 network; E.coli 1 inferred correlations above 50%; simulated E.coli 2 network; E.coli 2 inferred correlations above 50%; inferred networks made using 2 bins for each gene. Trisomic network Euploid network → Graph showing differences between Euploid and Trisomic Graph highlighting differences between Euploid and Trisomic using multiple datasets DREAM in-silico challenge Using phylogenetic profiles to predict protein function Basic Idea: Sequence alignment is a good way to infer protein function, when two proteins do the exact same thing in two different organisms. But can we decide if two proteins function in the same pathway? Assume that if the two proteins function together they must evolve in a correlated fashion: every organism that has a homolog of one of the proteins must also have a homolog of the other protein Phylogenetic Profile The phylogenetic profile of a protein is a string consisting of 0s and 1s, which represent the absence or presence of the protein in the corresponding sequenced genome; Protein P1: 0 0 1 0 1 1 0 0 Protein P2: 0 0 1 0 1 1 0 0 Protein P3: 1 0 0 1 0 1 0 0 For a given protein, BLAST against N sequenced genomes. If protein has a homolog in the organism n, set coordinate n to 1. Otherwise set it to 0. Phylogenetic Profile Species Proteins Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO, Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 96(8):4285-8,. 1999