* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Hardcastle, A., et. al. Pharmacodynamic markers of response to
History of molecular evolution wikipedia , lookup
List of types of proteins wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Interactome wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Protein adsorption wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Proteolysis wikipedia , lookup
Ligand binding assay wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Western blot wikipedia , lookup
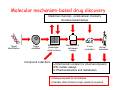
Pharmacodynamic markers of response to novel anticancer agents using a protein profiling sandwich immunoassay format Anthea Hardcastle, Matthew Cordwell, Peter Fong, Lindsay Stimson, Paul Workman and Wynne Aherne Cancer Research UK Centre for Cancer Therapeutics Institute of Cancer Research Sutton, Surrey, UK Molecular mechanism-based drug discovery Medicinal chemistry, combinatorial chemistry structure-based design Target Identification Target validation Biochemical, phenotypic, virtual screens Compound collections Mechanistic & cell-based assays In vivo evaluation Clinical evaluation Mechanistic endpoint or pharmacodynamic (PD) marker assays Pharmacokinetics and metabolism Assays required for all matrices Sample often limited so high sensitivity required Techniques for PD marker measurement Western blotting (Gold Standard’) Immunohistochemistry Flow cytometry Microplate immunoassays (ELISA) MSD Protein Profiling sandwich immunoassays ¾ Multiplex ‘catalogue’ assays ¾ Single assay development ‘in-house’ ¾ Option to multiplex ‘in-house’ and ‘catalogue’ assays The ease of validation for GCLP is an important consideration for PD assays as compliance with regulatory requirements is essential Meso Scale Discovery (MSD) technology Measured signal is light LIGHT z Can these assays be applied to different matrices? In vitro cell lysates Human tumour xenografts Clinical samples eg. PBMCs, tumours and plasma Aim : to carry out a feasibility study with inhibitors of HSP90 and HDAC PD markers for inhibitors of HSP90 The molecular chaperone HSP90 maintains the conformation, stability and function of oncogenic client proteins (eg. ERBB2, AKT and CDK4) HSP90 inhibitors cause degradation of client proteins, disruption of signalling pathways and antitumour activity Several agents currently in Phase I and II trials e.g. 17-AAG and 17-DMAG The molecular signature of HSP90 inhibition includes a fall in ERBB2, AKT and pERK and induction of HSP70 ‘Off the shelf’ MSD duplexed assays for PD markers of HSP90 inhibition 1 Phospho protein 2 SULFO-TAGTM labelled, detection antibody BSA A B BSA 1 2 3 4 Protein Capture Antibody Total Protein Working Electrode pERK Total ERK 35 Total ERBB2 25 20 15 10 1000 Ru/µg protein Ru/µg protein 30 750 500 5 250 0 0 DMSO Geld DMSO Geld HCT116 cells,1µM geldanamycin,24h. 1.25-10µg protein/well ‘Off the shelf’ duplexed assays for PD markers of HSP90 inhibition Total AKT pAKT 700 100 75 500 400 50 300 200 25 Phospho AKT Total AKT 600 100 0 0 DMSO Geld LY HCT116 cells,1µM Geldanamycin,20µM LY294002 (LY) 24h. 1.25-10µg protein/well ‘In-house’ MSD assay for HSP70 SULFO-TAGTM labelled, Goat anti rabbit IgG DELFIA HSP70 assay validated to Calibration curve Ru counts 100000 50000 0 0 1000 2000 3000 4000 5000 6000 7000 fmoles HSP70/well 2 Rabbit anti HSP70 A HSP70 Capture HSP70 Monoclonal Antibody B Working Electrode fmole HSP70/µg lysate GCLP In use for clinical trials of HSP90 inhibitors Assay transferred to MSD Potential for multiplexing HSP70 with client protein expression 1 1750 HCT116 ± 1 µM Geld 24h 1500 1250 1000 750 500 250 0 DMSO Geld PD markers for HDAC inhibitors HDACs catalyse the deacetylation of histones and are important for the regulation of gene expression HDACs are involved in the development and progression of malignancy HDACIs display antitumour activity and several compounds are being evaluated clinically Their precise mechanism of action is not clear but the most obvious result of HDAC inhibition is hyperacetylation of histones Hyperacetylation of histone H3 is used as a PD marker for HDAC inhibition ‘In-house’ assay for acetyl histone H3 (AcH3) SULFO-TAGTM labelled, Goat anti rabbit IgG Ac histone H3 Histones Capture Pan Histone monoclonal antibody Ru counts Rabbit anti AcH3 10000 7500 5000 Butyrate-treated HeLa cell extract 2500 0 Working Electrode Calibration curve 12500 0 100 150 200 250 300 ng/well AcH3 standard HCT116 5X GI50 SAHA 24h ng equivalents / µg lysate 50 DMSO SAHA 1000 AcH3 100 GAPDH 10 1 DMSO SAHA Assay being successfully used to measure the effect of novel HDACIs on AcH3 in human tumour xenografts Ex vivo treatment of PBMCs with HDACIs PBMCs and plasma separated from whole blood HDACIs SAHA and MS-275 or DMSO (control) added Incubated at 37°C for 4h PBMCs separated,washed and lysed 2 MS-275 AcH3 1 0 SAHA ng equivalents AcH3 3 Ex vivo PBMNCs 4h 5XGI50 DMSO AcH3 in lysates measured by MSD assay and western blot GAPDH DMSO SAHA MS275 This assay is being validated to GCLP for use in clinical trials of HDACIs Summary Experience with the MSD format is encouraging robust sensitive minimal matrix interference quantitative, higher-throughput alternative to western blots both catalogue and in-house assays used Provided proof of principle for mechanism of action in 2 therapeutic areas, HSP90 and HDAC, in different sample matrices Now used in a number of drug discovery projects in the Centre Assays are amenable to GCLP validation to comply with regulatory requirements for clinical trials Multiplexing potential for ‘in-house’ assays to be investigated Acknowledgements Matthew Cordwell, Peter Fong, Lindsay Stimson, Paul Workman and Wynne Aherne Colleagues in the Cancer Research UK Centre for Cancer Therapeutics Members of the Analytical Technology and Screening Team Tuc Ahmad from MSD