* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Using bacterial biosensors to understand the genetic basis for
Quantitative trait locus wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genetic engineering wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Oncogenomics wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Transposable element wikipedia , lookup
Ridge (biology) wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Pathogenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Microevolution wikipedia , lookup
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Public health genomics wikipedia , lookup
Genome evolution wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
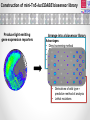
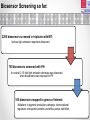
Using Pseudomonas aeruginosa bioluminescent biosensors to understand the genetic basis for antimicrobial resistance Angharad Ellen Green1 ([email protected]) Eshwar Mahenthiralingam1, Thomas Connor1, Yvan Le Marc2 and Alejandro Amézquita2 1Cardiff 2Unilever, School of Biosciences, Cardiff University, Cardiff, Wales, UK. Safety and Environment Assurance Centre (SEAC), Bedfordshire, England, UK. Antibiotic Resistance Mechanisms Workshop for researchers 27th November 2015 Presentation overview 1. Pseudomonas aeruginosa as a contaminant of industrial products Introduction 2. Use of preservatives in home and personal care products 3. Developing bacterial biosensors and exposing them to industrial preservatives Project Aims 4. Identifying differentially regulated genes after preservative exposure 5. Optimising industrial preservative formulations Project outcomes & conclusions Pseudomonas aeruginosa • • Gram-negative opportunistic pathogen Important clinical microorganism • Successful contaminant of home and personal care products • Metabolically and genetically versatile • Ability to survive with minimal nutritional requirements Large 5.2-7.0 Mbp genome • High tolerance of antimicrobials Limited knowledge of industrially isolated populations of P. aeruginosa Industrial Preservatives • Class of biocide with a broad range of activity • Globally, the most widely used antimicrobial by weight • Added to home and personal care products to control microbial growth • P. aeruginosa is able to survive in the presence of industrial preservatives • Mechanisms of resistance specific to preservatives are not well characterised Antibiotics Vs. Preservatives Preservatives Antibiotics Vs. Lewis, (2013) Limited number of known resistance mechanisms specific to preservatives Arias and Murray, (2012) Project Overview Aim: Map the genetic and metabolic pathways associated with Pseudomonas aeruginosa preservative insusceptibility Generate an industrial Pseudomonas bioluminescent mini-Tn5-luxCDABE biosensor library Identify differentially expressed genes on exposure to industry relevant preservatives Predict bacterial responses to industrial preservative formulations Bacterial Bioluminescent Biosensors Construction of mini-Tn5-luxCDABE biosensor library Industrially isolated Pseudomonas bacteria Light emitting reporter transposon inserts randomly within the genes (Winson et al, 1998) Construction of mini-Tn5-luxCDABE biosensor library Produce light emitting gene expression reporters Arrange into a biosensor library Advantages: • Direct screening method • Reusable • Low cost • Assess response of numerous genes to a variety of antimicrobials Disadvantages: • Luciferase inactivation • Derivatives of wild type = predictive method of analysis • Lethal mutations Screening preservatives using bacterial biosensors Biosensors libraries are exposed to preservatives Screening preservatives using bacterial biosensors Identify biosensors with a change in light emission Biosensor of interest Preservative responding biosensor 24 hour exposure to the industrial preservative Benzisothiazolinone (BIT): Pigment overproducer Preservative responding biosensor 24 hour growth on plates containing the industrial preservative BIT: Light emission increase Preservative responding biosensor 24 hour exposure to the industrial preservative Phenoxyethanol (PH): Preservative responding biosensor 24 hour growth on plates containing the industrial preservative PH: Light emission Decrease Identifying differentially regulated genes after preservative exposure Identifying transposon insertion sites 1. Isolation of biosensor genomic DNA 2. Digestion 3. Ligation of digested product 4. Ligated product amplified by inverse PCR 5. PCR products sequenced with nested primers x10-6 PCR product amplification 6. Sequence mapped back to the wild type strain/ Pseudomonas genome database Identifying transposon insertion sites Identifying transposon insertion sites Biosensor Screening so far: 2200 biosensors screened in triplicate with BIT: Various light emission responses observed 750 biosensors screened with PH: An overall 2.15 fold light emission decrease was observed when biosensors were exposed to PH 100 biosensors mapped to genes of interest: Mutations in pigment production pathways, transcriptional regulators, transporter proteins and efflux pumps identified Project Outcomes and Conclusions Project Outcome: Optimising preservative formulations Responding biosensors linked to metabolic networks and gene pathways will be used to predict responses to various combinations of preservatives Conclusions Project has so far demonstrated: • Successful construction of a light emitting biosensor library using an industrially isolated strain of Pseudomonas aeruginosa. • Screening using sub-MIC preservatives identified biosensors with altered light expression in both planktonic and agar growth. • Inverse PCR successfully identified transposon insertion sites; differential gene expression will be validated by real-time PCR. Research Outcomes and Next Steps: • Continue screening to develop a biosensor panel for optimising preservative formulations. • Global gene expression to preservatives will also be mapped by RNA-seq. Acknowledgements • Cardiff School of Biosciences, Cardiff University Supervisors: Professor Eshwar Mahenthiralingam and Dr. Thomas Connor. Lab Team: Dr. Matt Bull, Dr. Beky Weiser, Dr. Laura Rushton, Dr. Cerith Jones and Rachel Rowe. • Unilever, Safety and Environment Assurance Centre (SEAC) Dr. Yvan Le Marc and Dr. Alejandro Amézquita. Any questions?