* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download The postsynaptic NMDA-receptor–PSD-95
Index of biochemistry articles wikipedia , lookup
Protein adsorption wikipedia , lookup
Gene regulatory network wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Western blot wikipedia , lookup
Biochemical cascade wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Cell membrane wikipedia , lookup
Neurotransmitter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Endomembrane system wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
NMDA receptor wikipedia , lookup
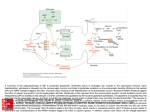
CELL SCIENCE AT A GLANCE The postsynaptic NMDA-receptor–PSD-95 signaling complex in excitatory synapses of the brain Morgan Sheng MGH and Harvard Medical School, Howard Hughes Medical Institute (Wellman 423), Massachusetts General Hospital, 50 Blossom Street, Boston, MA 02114, USA Excitatory synapses of the brain use as neurotransmitter the amino acid glutamate, which is released in packets from the presynaptic terminal. The postsynaptic membrane is specialized for the reception of glutamate signals and the transduction of these signals into the postsynaptic cell. Containing a high concentration of glutamate receptors and associated cytoskeletal and signaling proteins, the postsynaptic specialization is visible by electron microscopy as a thickening (~30 nm thick) of the postsynaptic membrane known as the Ca2+ 1251 postsynaptic density (PSD). The NMDA subtype of glutamate receptor (NMDAR) is an abundant constituent of the PSD and functions as a glutamategated calcium-permeable ion channel. NMDARs are anchored in the PSD by interactions between the cytoplasmic Cterminal tails of their NR2 subunits and the PDZ domains (orange) of PSD-95, an abundant PSD protein that forms a two-dimensional lattice immediately under the postsynaptic membrane. The PDZ domains of PSD-95 also bind to other postsynaptic membrane proteins, including potassium channels (K+ Ch), tyrosine kinases (ErbB4) and cell adhesion molecules (neuroligin). Having multiple domains that bind to a variety of cytoplasmic proteins, PSD-95 functions as a scaffold to assemble a specific set of signaling proteins around the NMDAR. These proteins, such as neuronal nitric oxide synthase (nNOS), SynGAP [a GTPase-activating protein (GAP) for Ras] and SPAR (a GAP for Rap), may participate in downstream signaling by NMDARs. PSD-95, in turn, bB B4 interacts with GKAP and Shank, two scaffold proteins lying in the deep part of the PSD. Shank binds to Homer, which interacts directly with the cytoplasmic tail of the metabotropic glutamate receptor (mGluR), a Gprotein-coupled glutamate receptor. Homer forms multimers and additionally binds to the inositol 1,4,5-trisphosphate receptor [Ins(1,4,5)P3R] found in smooth endoplasmic reticulum (SER), thereby linking cell surface mGluRs to a downstream effector [Ins(1,4,5)P3R] in intracellular calcium stores. Thus an extensive network of protein-protein interactions within the PSD links together different classes of postsynaptic glutamate receptor and couples them to specific intracellular signaling pathways. Cell Science at a Glance on the Web Electronic copies of the full-size poster insert are available in the online version of this article (see www.biologists.com/jcs). Files in several formats are provided and may be downloaded for use as slides. Neuroligi u in N NMDAR CaMKII GKAP P 2+ 2 + (See poster insert) 1252 JOURNAL OF CELL SCIENCE 114 (7) Year 2001 Travelling Fellowships JCS offers fellowships of up to US$4000 to graduate students and post-docs wishing to make collaborative visits to other laboratories. These are designed to cover the cost of travel and other expenses, and there is no restriction on nationality. Applicants should be working in the field of cell biology and intend to visit a laboratory in another country. Each application is judged on the excellence of the candidate, and the importance and innovative quality of the work to be done. Application forms can be downloaded from our Web site at www.biologists.com/cob/tf. Please send the completed application form, together with a copy of your CV, an account of the work to be done and a breakdown of the costs involved, as well as letters of recommendation from the heads of the laboratory in which you currently work and the laboratory you hope to visit, to the Production Editor at the address below. Journal of Cell Science Editorial Office The Company of Biologists Limited Bidder Building 140 Cowley Road Cambridge CB4 0DL, UK Deadline: 30 June 2001